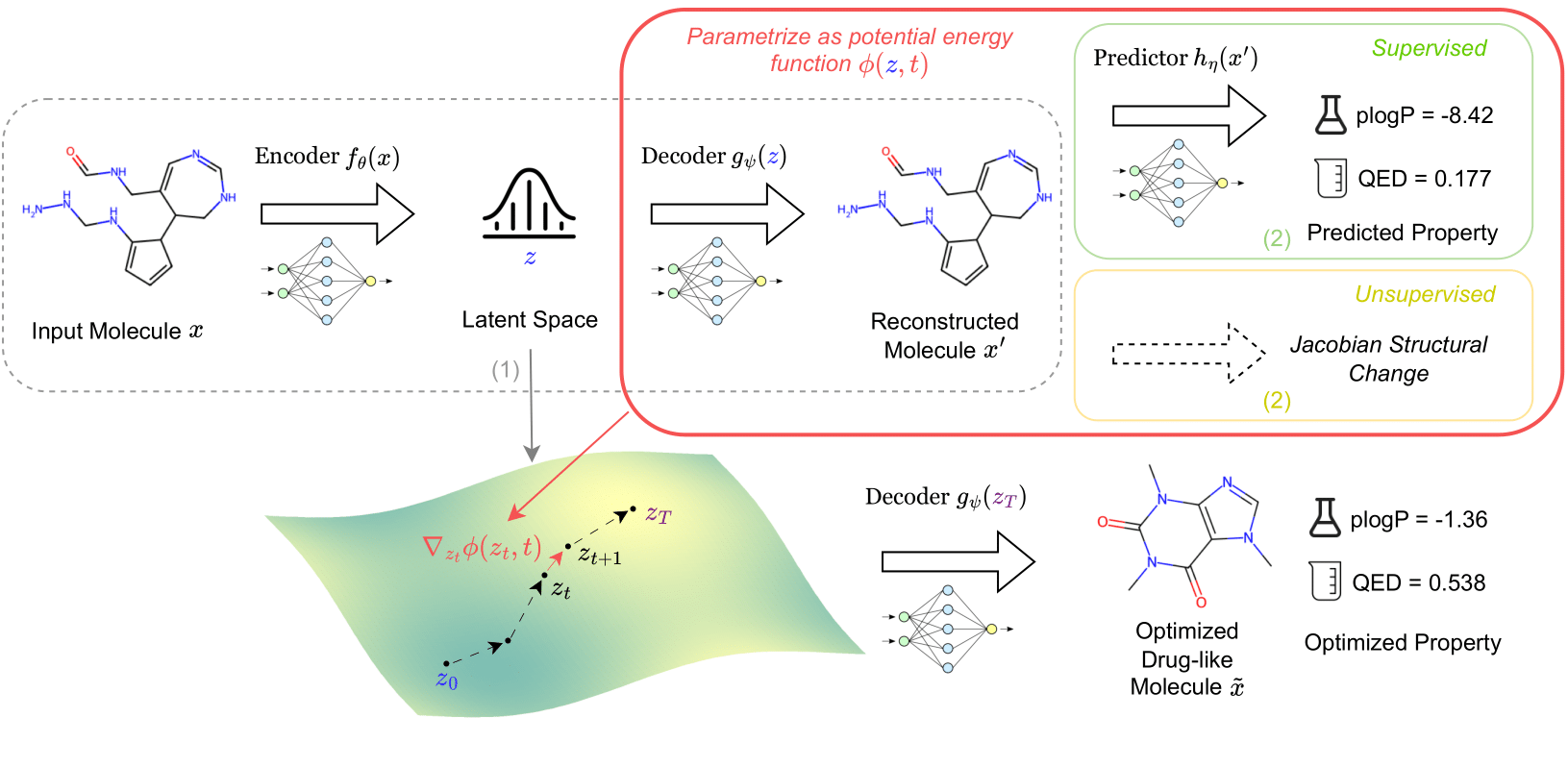
This repo implements the paper 🔗: Navigating Chemical Space with Latent Flows by Guanghao Wei*, Yining Huang*, Chenru Duan, Yue Song, and Yuanqi Du.
Flows can uncover meaningful structures of latent spaces learned by generative models! We propose a unifying framework to characterize latent structures by flows/diffusions for optimization and traversal.
Try our live demo here!
- Install all dependencies with
conda env create -f environment.yml.- (Optional) Install AutoDock-GPU for docking binding affinity. See Notes on Compiling AutoDock-GPU.
- (Recommended)
mv .env.defaults .envand specifyPROJECT_PATHin.env. This is later used to run the experiments in the project root directory.
- Download data and put it in the
datadirectory. - Train the VAE model by running
python experiments/train_vae.py.- (Optional) Download our pre-trained VAE model checkpoint, see Download Data & Model Checkpoints.
- For supervised learning
- Prepare the data by running
python experiments/prepare_random_data.py. - Train the supervised surrogate predictor by running
bash experiments/supervised/train_prop_predictor.sh. - Train the energy network with supervised semantic guidance by
running
bash experiments/supervised/train_wavepde_prop.sh.
- Prepare the data by running
- For unsupervised learning
- Train the energy network with unsupervised diversity guidance by running
python experiments/train_wavepde.py. - Compute the pearson correlation coefficient by running
python experiments/unsupervised/corr.py. Refer tonotebooks/experiments/unsupervised/corr.ipynbfor more details. - Modify
experiments/utils/traversal_step.pyin place with the best correlation coefficient index.
- Train the energy network with unsupervised diversity guidance by running
- To reproduce the experiment results from the paper, run the following commands:
bash experiments/optimization/optimization.shfor similarity constrained optimization.bash experiments/optimization/uc_optim.shfor unconstrained optimization.python experiments/optimization/optimization_multi.pyfor multi-objective optimization.bash experiments/success_rate/success_rate.shfor molecule manipulation tasks.
We used lightning(doc)
and tap(doc) to parse the arguments.
Following is an example command to pass in arguments configured by lightning:
python experiments/supervised/train_prop_predictor.py \
-e 50 \
--model.optimizer sgd \
--data.n 11000 \
--data.batch_size 100 \
--data.binding_affinity true \
--data.prop 1errWe extracted 4,253,577 molecules from the three commonly used datasets for drug discovery including MOSES, ZINC250K(download), and ChEMBL.
- The processed dataset and VAE model checkpoints are available
at Google Drive.
- Data processing notebooks refers to
notebooks/datasets.ipynb.
- Data processing notebooks refers to
Notes on Compiling AutoDock-GPU
The conda version of AutoDock-GPU is not compatible with RTX 3080 & 3090.
So don't use environment.yml to install AutoDock-GPU.
Make sure to follow this issue to
compile the source code.
A good reference for the SM code
can be found here.
Some commands might be useful:
export GPU_INCLUDE_PATH=/usr/local/cuda/include
export GPU_LIBRARY_PATH=/usr/local/cuda/lib64
make DEVICE=CUDA NUMWI=128 TARGETS=86To test if the compilation is successful, run the following command:
obabel -:"CCN(CCCCl)OC1=CC2=C(Cl)C1C3=C2CCCO3" -O demo.pdbqt -p 7.4 --partialcharge gasteiger --gen3d
autodock_gpu_128wi -M data/raw/1err/1err.maps.fld -L demo.pdbqt -s 0 -N demo@misc{wei2024navigating,
title={Navigating Chemical Space with Latent Flows},
author={Guanghao Wei and Yining Huang and Chenru Duan and Yue Song and Yuanqi Du},
year={2024},
eprint={2405.03987},
archivePrefix={arXiv},
primaryClass={cs.LG}
}