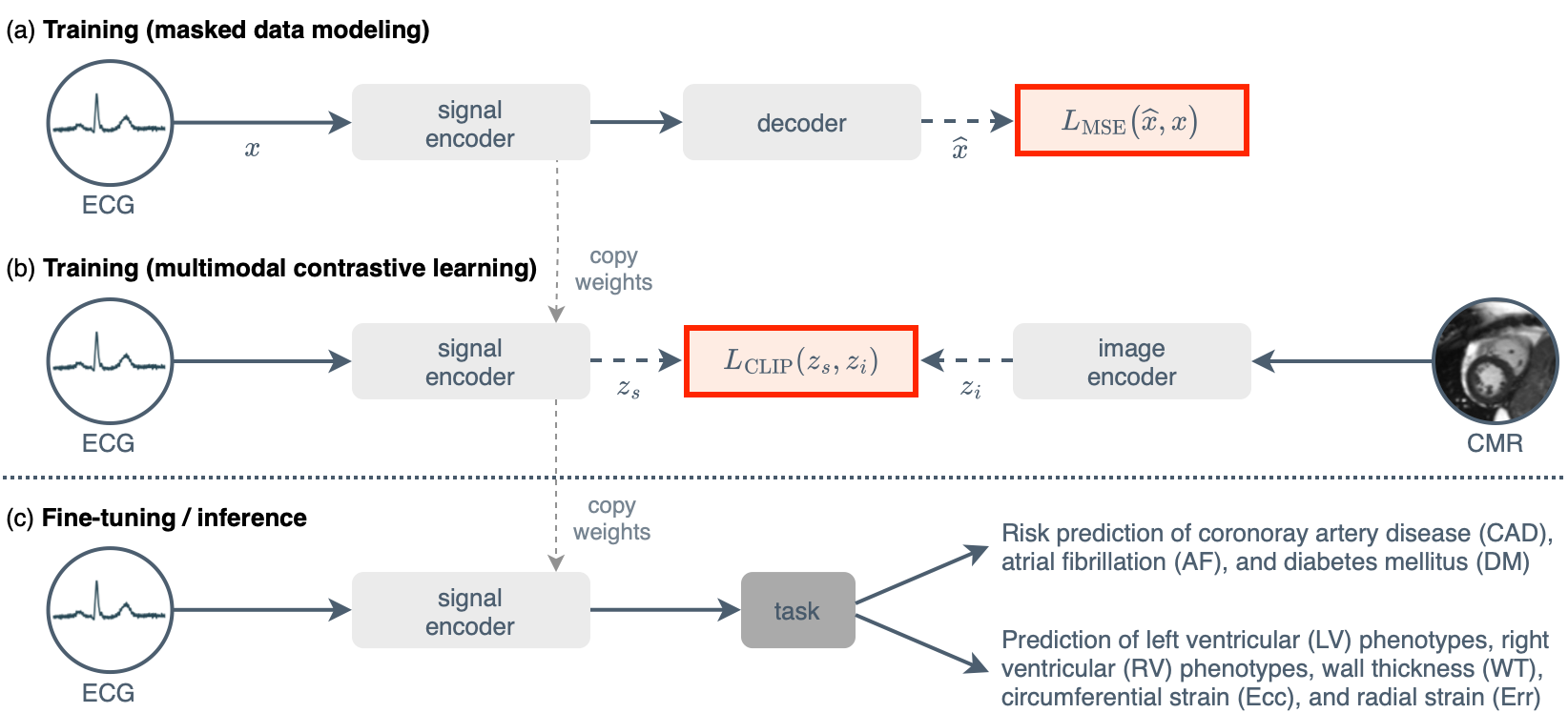
This is the official implementation of our paper Unlocking the Diagnostic Potential of ECG through Knowledge Transfer from Cardiac MRI (2023).
If you find the code useful, please cite
@article{turgut2023unlocking,
title={Unlocking the Diagnostic Potential of ECG through Knowledge Transfer from Cardiac MRI},
author={Turgut, {\"O}zg{\"u}n and M{\"u}ller, Philip and Hager, Paul and Shit, Suprosanna and Starck, Sophie and Menten, Martin J and Martens, Eimo and Rueckert, Daniel},
journal={arXiv preprint arXiv:2308.05764},
year={2023}
}
Install environment using conda env create --file environments/mae.yaml.
Install timm library using pip install -e pytorch-image-models.
For detailed instructions to run the code for unimodal pre-training, see the PRETRAIN.md of the mae subfolder.
Install environment using conda env create --file environments/mmcl.yaml.
Install timm library using pip install -e pytorch-image-models.
For detailed instructions to run the code for multimodal pre-training, see the README.md of the mmcl subfolder.
Install environment using conda env create --file environments/mae.yaml.
Install timm library using pip install -e pytorch-image-models.
For detailed instructions to run the code for fine-tuning and inference, see the FINETUNE.md of the mae subfolder.