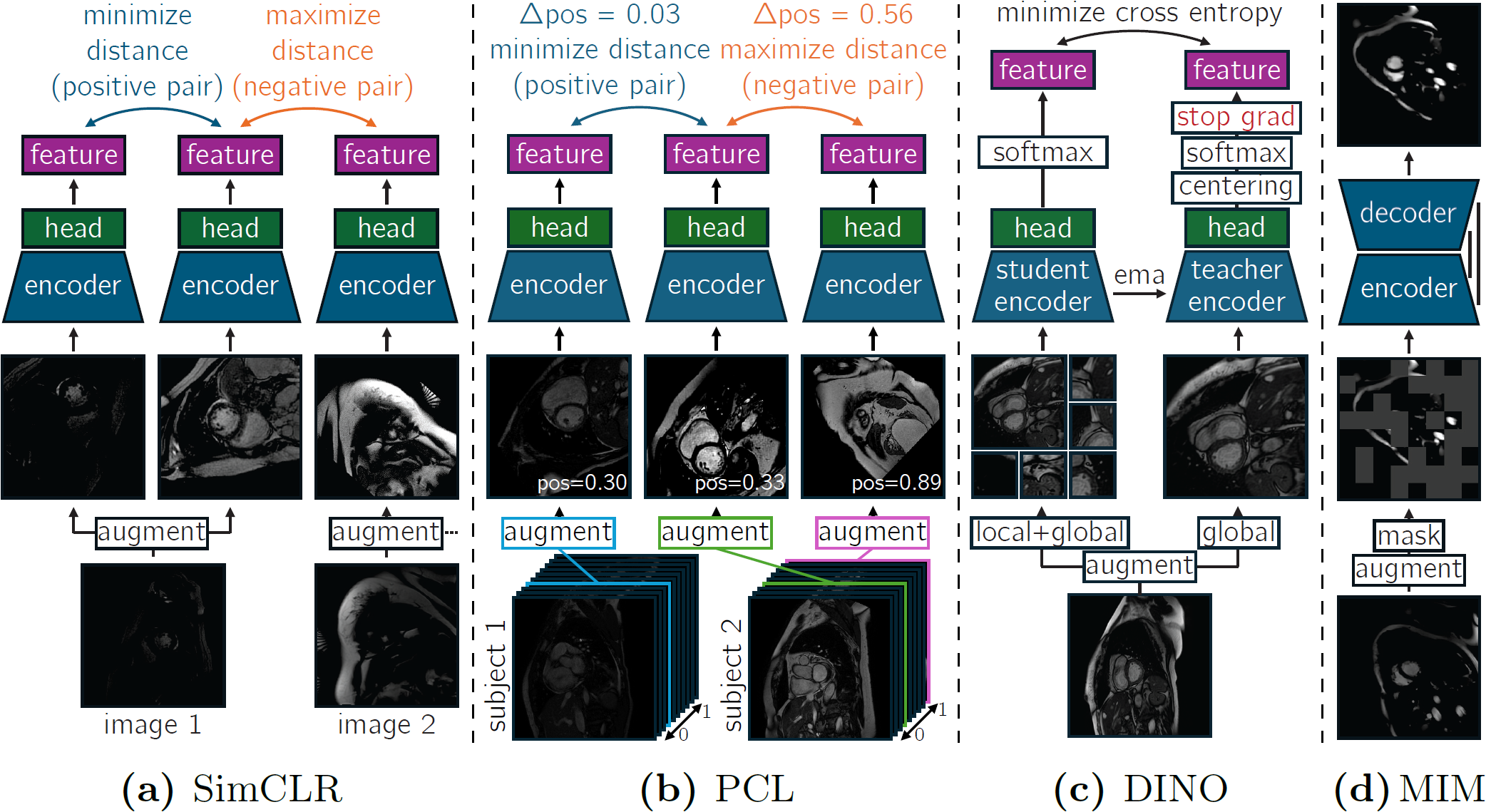
PyTorch based code used for Self-supervised pretraining for cardiovascular magnetic resonance cine segmentation, comparing 4 self-supervised pretraining (SSP) methods and an nnU-Net based baseline for 2D cardiovascular magnetic resonance cine segmentation. The 4 methods are: SimCLR, positional contrastive learning (PCL), DINO, and masked image modeling (MIM), which are all visualized below:
The code uses our own qcardia-data and qcardia-models modules.
It is recommended to make a new environment (tested for Python 3.11.9) and first installing PyTorch and checking GPU availability. Installation instructions for the latest stable PyTorch version can be found in their "get started" guide. Alternatively, they include instructions for previous stable versions. Installing the PyTorch version the package was tested for (PyTorch 2.3.1) might limit warnings or unexpected behaviours.
The requirements.txt file lists the required packages, mostly by installing our own qcardia-data and qcardia-models modules. It can be installed using pip:
pip install -r /path/to/requirements.txt
Alternatively, local (editable) copies of qcardia-data and qcardia-models can be used. Installation instructions for local (editable) copies can be found on the respective GitHub pages of the qcardia packages.
Public datasets must first be reformatted so the qcardia-data pipeline can use the data. For the supported public datasets, this can be achieved by:
- Downloading the public data.
- Saving the data with the expected folder hierarchy.
- Updating the configs to point to your local data folder.
- Running the relevant data setup functions.
The public M&Ms and M&Ms-2 challenge datasets can be downloaded from their respective websites. After downloading, our automatic qcardia-data reformatting can be used when you've saved the original datasets in their expected folder heirarchy:
data
└── original_data
├── MnM
│ ├── dataset
│ │ ├── A0S9V9
│ │ │ ├── A0S9V9_sa_gt.nii.gz
│ │ │ └── A0S9V9_sa.nii.gz
│ │ └── ...
│ └── 211230_M&Ms_Dataset_information_diagnosis_opendataset.csv
├── MnM2
│ ├── dataset
│ │ ├── 001
│ │ │ ├── 001_SA_CINE.nii.gz
│ │ │ ├── 001_SA_ED_gt.nii.gz
│ │ │ ├── 001_SA_ED.nii.gz
│ │ │ ├── 001_SA_ES_gt.nii.gz
│ │ │ ├── 001_SA_ES.nii.gz
│ │ │ └── ... (001_LA... -> long axis data unused for now)
│ │ └── ...
│ └── dataset_information.csv
└── ...
The qcardia-data cine setup function can be then be used, requiring only a path to your local data folder:
from qcardia_data.setup import setup_cine
from pathlib import Path
data_path = Path("path/to/your/data_folder")
setup_cine(data_path)The data setup functions reformat the relevant available original datasets, and can generate default test data splits. A copy of the test split file, as well as a full data split file, are included in this repository. By default, the included configs use the full split file, which should be saved in the subject_splits subfolder in your data folder. Also make sure to update your data path in any config files.
More details can be found in the qcardia-data demo and README.
Afterwards, the training scripts should work out of the box. The train_unet script trains a U-Net based on the baseline-config configuration file. Similarly, there are training scripts for SimCLR, PCL, DINO, and MIM pretraining, using their respective config files. Note that the data path should be updated in each config file.
If you find our work useful in your research please consider citing our paper:
@misc{demooij2024ssp,
title={Self-supervised Pretraining for Cardiovascular Magnetic Resonance Cine Segmentation,
author={Rob A. J. de Mooij and Josien P. W. Pluim and Cian M. Scannell},
year={2024},
eprint={2409.18100},
archivePrefix={arXiv},
primaryClass={cs.CV},
url={https://arxiv.org/abs/2409.18100},
}