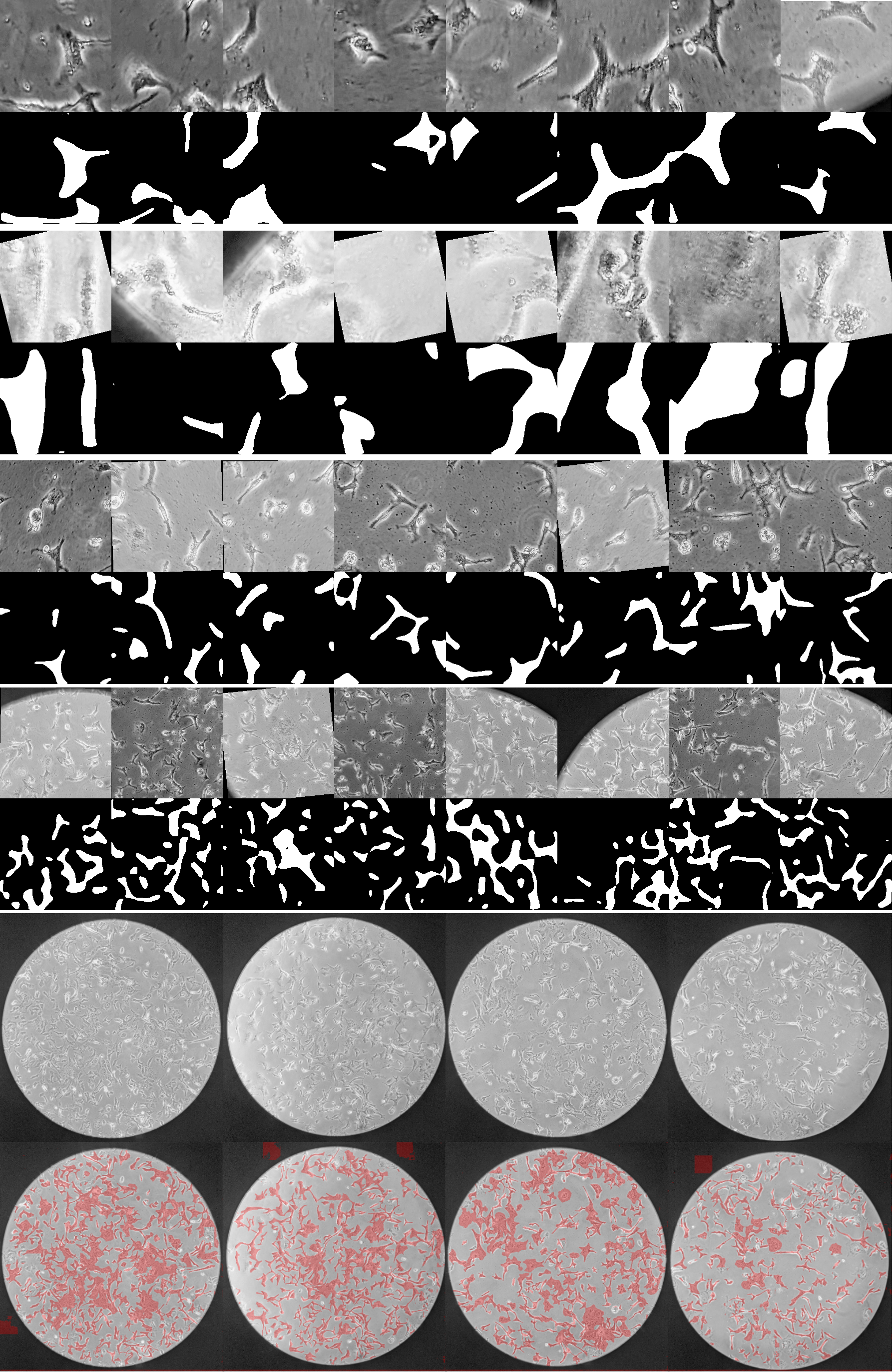
This repository holds code for stem cell segmentation. The codes are built based on the original TransUNet repo: TransUNet.
Our NT2 cell dataset is available at https://doi.org/10.15131/shef.data.25604421
Please prepare an environment with python=3.10, We recommand using Anaconda3.
After installing python(Anaconda), please run the following command for installing the dependencies.
pip install -r requirements.txtTo train your own model from zero please download the Google ViT pre-trained models
- Get models in this link: R50-ViT-B_16, ViT-B_16, ViT-L_16... or run the following commands replacing the {MODEL_NAME} to one of the R50-ViT-B_16, ViT-B_16, ViT-L_16..
wget https://storage.googleapis.com/vit_models/imagenet21k/{MODEL_NAME}.npz &&
mkdir ../model/vit_checkpoint/imagenet21k &&
mv {MODEL_NAME}.npz ../model/vit_checkpoint/imagenet21k/{MODEL_NAME}.npz*For inference only, download the provided checkpoint, you will need to download the whole folder and put it under the ./model/ path. Create model folder if you dont have one.
Prepare the data and generate Train and Test Lists:
Download the data first, then run the following script to spilit the data into training and testing 224x224 patches, you will need to change the data path in the script:
python data_pre.py --mode train_test --divide --image_dir /path/to/images --mask_dir /path/to/masks --dataset_dir /path/to/training_and_testing_patches --lists_dir /path/to/listsFor multi images inference only: Generate Eval List by running:
python data_pre.py --mode eval --image_dir /path/to/images --lists_dir /path/to/listsThe whole data and the lists should be under a directory structure like this:
Cellseg_UOS/
├── data/
│ └── Cell/
│ ├── image1.png
│ ├── image1_mask.png
│ ├── image1_ps224_0_0.png
│ ├── image1_ps224_0_0_mask.png
│ ├── image1_ps448_0_0.png
│ ├── image1_ps448_0_0_mask.png
│ ├── ...
│ ├── image2.png
│ ├── image2_mask.png
│ ├── image2_ps224_0_0.png
│ ├── image2_ps224_0_0_mask.png
│ ├── image2_ps448_0_0.png
│ ├── image2_ps448_0_0_mask.png
│ ├── ...
├── model/
│ ├── vit_checkpoint/
│ │ └── imagenet21k/
│ │ ├── R50+ViT-B_16.npz # Pretrained model checkpoint
│ │ └── *.npz # Other model checkpoints
│ └── TU_CellSeg224/
│ └── TU_pretrain_R50-ViT-B_16_skip0_epo200_bs128_224_St150_SN100_SEL10000_SF100000_LTpareto_VOS/
│ ├── epoch_199.pth # Model weights
│ └── ...
└── lists/
├── train.txt
├── test.txt
├── eval.txt
├── datasets/
├── vis/
├── data_pre.py
├── train.py
├── test.py
├── ...Example to train the model with outlier argument:
python train.py --batch_size 256 --max_epochs 200 --vit_name ViT-B_16 --start_epoch 150 --select 10000 --sample_from 100000 --loss_type norm --use_vos- Run the test with the same configs on the test set:
python test.py --batch_size 128 --max_epochs 200 --vit_name R50-ViT-B_16 --start_epoch 150 --select 10000 --sample_from 100000 --loss_type $loss_type --use_vosVisualize the high resolution raw images using the model trained by:
python test.py --volume_path 'your path to images' --max_epochs 200 --batch_size 256 --vit_name R50-ViT-B_16 --is_savenii --data_split eval --start_epoch 150 --select 10000 --sample_from 100000 --loss_type norm --use_vosFor single image visualization, please open the inference.ipynb in jupyter notebook, put your testing images under the folder ./vis/, and then run the notebook for results. Note that you still need to install the dependencies first (Step 1).
Please note that if you are not using the provided checkpoint, you will need to change the snapshot_path and other configs in the notebook.