Manipulate and draw trees in Newick format.
Binary files are self-contained, no dependencies.
Linux newick
Windows newick.exe
Make a subset tree given file with leaf labels, one per line (labels
do not need to be a subtree, the tree is collapsed as needed):
newick -input tree.newick -labels labels.txt -output subset.newick
Get leaf labels:
newick -getlabels tree.newick -output labels.txt
Report miscellaneous information about a Newick file:
newick -stats trees.newick
Calculate Robinson-Foulds (R-F) distance between two trees:
newick -rofo tree1.newick -tree2 tree2.newick -log rofo.log
Calculate all-vs-all R-F distances between trees in Newick file:
newick -rofos trees.newick -log rofos.log
Re-label trees, labels.tsv tab-separated with #1=old_label #2=new_label:
newick -relabel trees.newick -labels labels.tsv -output relabeled_trees.newick
Add integer node number labels to internal nodes:
newick -intlabel tree.newick -output intlabel.newick
Root by outgroup, specify labels.txt with leaf labels of outgroup or GroupName which
is a substring of the outgroup labels, e.g. phylum name if format is A1234.Phylum:
newick trees.newick [-labels labels.txt | -outgroup GroupName] -output rooted.newick
Convert tab-separated to Newick:
newick -tsv2newick tree.tsv -output tree.newick
Convert Newick to tab-separated:
newick -newick2tsv tree.newick -output tree.tsv
Ladderize trees by rotating internal nodes so that larger subtree is always the
left (default) or right subtree:
newick -ladderize trees.newick -output ladderized.newick [-right]
Split tree into N roughly equal-sized subtrees (clusters), output is N files
named prefixi, i=1..N containing labels for each subtree:
newick -split tree.newick -n N -prefix prefix
Convert trees to cladograms (leaves equidistant from root):
newick -clado trees.newick -output clado.newick
Calculate edge confidence values from set of bootstrapped trees:
newick -conf tree.newick -trees replicates.newick -output conftree.newick
Condense a tree by identifying best-fit nodes for each feature group and making
a tree of just those nodes; unary edges are collapsed by summing lengths and
taking max confidence, leaves are labeled with features (e.g. phylum names):
newick -condense trees.newick -features features.tsv -output condensed.newick
Extract just the branching order by collapsing unary nodes, deleting all edge lengths
and deleting all confidence values (all internal node labels removed):
newick -topo trees.newick -output topos.newick
Delete one or more leaves and collapse any resulting unary nodes, useful e.g. for
deleting outgroup to simplify figure:
newick -deleteleaves trees.newick [-label OutgroupName | -labels labels.txt] -output .newick
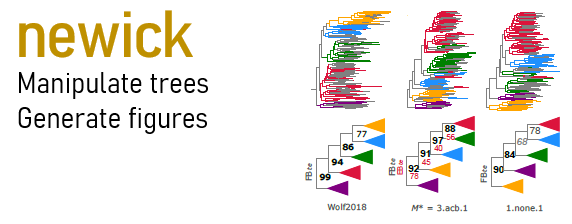
Draw one tree or several trees with optional coloring of edges:
newick -draw tree.newick -svg figure.svg
newick -drawf tree.newick -features features.tsv -colors colors.tsv -svg figure.svg
newick -drawfs trees.newick -features features.tsv -colors colors.tsv -svg figure.svg
-features is tsv file with #1 leaf_label #2 feature_name (e.g. phylum).
-colors is tsv file with #1 feature_name #2 color, where color is any valid svg color,
can be rgb, hex or name e.g. red.
-default_color color
Color for unlabeled edges (default gray).
-title text
Title text.
-title_font_size n
Title font size (default 10).
-unitlengths
Treat all edge lengths as 1 (phylogram).
-strokewidth n
Line width for edges (default 1).
-tree_width n
Width of tree (default 1000).
-tree_height n
Height of tree (default 1000).
-tree_spacing n
Space between trees (default 300).
-trees_per_row n
Number of trees per row in figure (default 4).
-triangles w,h
Draw triangles at leaves with width w and height h.
-legend legend.svg
Legend showing features (e.g. phylum names) and colors.