The following software is required:
- Apptainer (tested with version 1.2.4)
- Nextflow (tested with version 23)
- Only when using
enable_summary = true. Python 3 (tested with version 3.7.7) with the following packages: pysam, nbconvert, ipykernel, pandas.
Caution
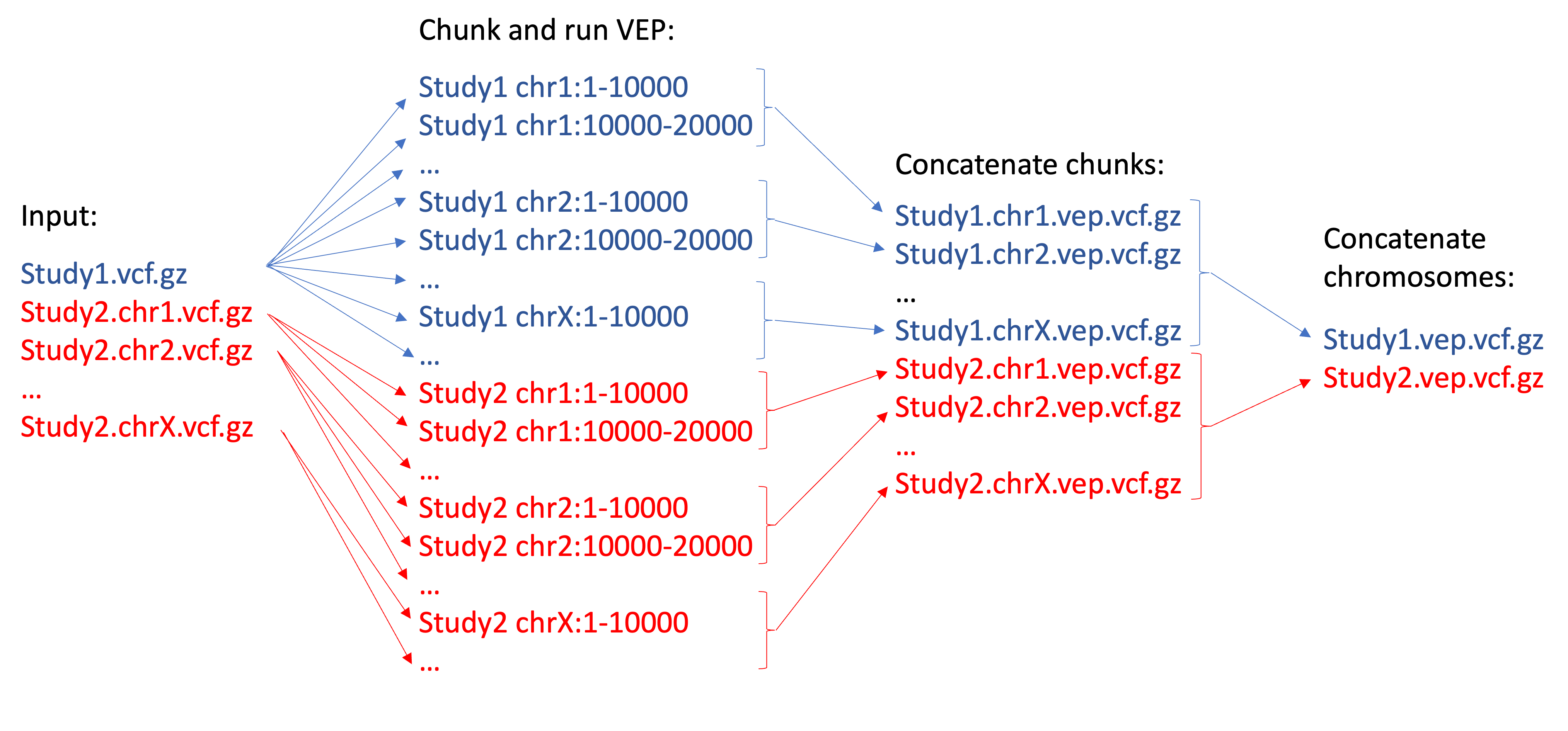
- Your input VCFs must be indexed and have corresponding
.tbifiles - Input VCFs from the same study must have the same prefix, which can't be a chromosome name.
- Input VCFs can be split by chromosome as long as they all have the same prefix e.g., [prefix].chr1, [prefix].chr2, ..., [prefix].chrY.
Note
If you are a member of the CERC-Genomic-Medicine team, then all necessary files to run VEP are already installed on the server. Consult with your colleagues on where to find them.
This section describes how to set up VEP, download all necessary cache files, and install LoFtee plugin.
-
Load
apptainermodule:module load apptainer -
Build the SIF image with additional tools:
samtools,bcftools, andDBD::SQLite.Use SylabsCloud free Remote Builder service to create the SIF image remotely. Specify the following in your definition file (i.e.
.deffile):Bootstrap: docker From: ensemblorg/ensembl-vep:latest %post apt-get update -y apt-get install -y samtools apt-get install -y bcftools apt-get install -y libdbd-sqlite3-perlAfter you built the SIF image using the Remote Builder web interface, pull the image to the cluster.
Confirm that the
SylabsCloudremote service is available (if not, you will need to add it):apptainer remote listLogin to the SylabsCloud remote service
apptainer remote login SylabsCloudPull the SIF image using the repository name you speficied when creating the SIF image:
apptainer pull vep.sif library://dtaliun/remote-builds/vep:23may2024 -
Download VEP cache files into local
vep_cachedirectory:mkdir `pwd`/vep_cache export CURL_CA_BUNDLE=/etc/ssl/certs/ca-certificates.crt apptainer run -B `pwd`/vep_cache:/opt/vep/.vep vep.sif INSTALL.pl -a cf -s homo_sapiens -y GRCh38 -c /opt/vep/.vepThis step may take more than 1h.
More detailed instructions on how to set up LoFtee are here.
-
You must clone LoFtee repository into your local
vep_cachedirectory:cd vep_cache git clone https://github.com/konradjk/loftee.git loftee_GRCh37 git clone https://github.com/konradjk/loftee.git loftee_GRCh38 cd loftee_GRCh38 git checkout grch38 cd .. -
Download all necessary databases (based on human genome build you plan to use) as described here into your
vep_cachedirectory into foldersloftee_db_GRCh37andloftee_db_GRCh38. These should include: GERP conservation scores (only for GRCh38), human_ancestor.fa files, SQL databases with PhyloCSF metrics (SQL files must be unzipped).
- Download VEP plugins into
vep_cachedirectory:cd vep_cache git clone https://github.com/Ensembl/VEP_plugins.git Plugins cd .. - Download CADD scores for GRCh37 and GRCh38 builds
cd vep_cache mkdir CADD_GRCh37 cd CADD_GRCh37 wget https://krishna.gs.washington.edu/download/CADD/v1.7/GRCh37/whole_genome_SNVs.tsv.gz wget https://krishna.gs.washington.edu/download/CADD/v1.7/GRCh37/whole_genome_SNVs.tsv.gz.tbi wget https://krishna.gs.washington.edu/download/CADD/v1.7/GRCh37/gnomad.genomes-exomes.r4.0.indel.tsv.gz wget https://krishna.gs.washington.edu/download/CADD/v1.7/GRCh37/gnomad.genomes-exomes.r4.0.indel.tsv.gz.tbi cd .. mkdir CADD_GRCh38 cd CADD_GRCh38 wget https://krishna.gs.washington.edu/download/CADD/v1.7/GRCh38/whole_genome_SNVs.tsv.gz wget https://krishna.gs.washington.edu/download/CADD/v1.7/GRCh38/whole_genome_SNVs.tsv.gz.tbi wget https://krishna.gs.washington.edu/download/CADD/v1.7/GRCh38/gnomad.genomes.r4.0.indel.tsv.gz wget https://krishna.gs.washington.edu/download/CADD/v1.7/GRCh38/gnomad.genomes.r4.0.indel.tsv.gz.tbi cd .. cd ..
- Copy
Plugins/CONTEXT.pmfile from this repository to thevep_cache/Pluginsdirectory.cd Plungins wget https://raw.githubusercontent.com/CERC-Genomic-Medicine/vep_pipeline/master/Plugins/CONTEXT.pm cd ..
After above steps, your local vep_cache directory should be similar to this:
|- vep_cache
|- homo_sapiens (directory with VEP databases)
|- loftee_GRCh37 (loftee scripts for build GRCh37)
|- loftee_GRCh38 (loftee scripts for build GRCh38)
|- loftee_db_GRCh37 (loftee databases for build GRCh37)
|- loftee_db_GRCh38 (loftee databases for build GRCh38)
|- Plugins (CADD plugin)
|- CADD_GRCh37 (CADD scores for build GRCh37)
|- CADD_GRCh38 (CADD scores for build GRCh38)
-
Clone this repository to the directory where you will run the pipeline:
git clone https://github.com/CERC-Genomic-Medicine/vep_pipeline.git -
Modify
nextflow.configconfiguration file.params.vcfs-- path to your VCF/BCF file(s). You can useglobexpressions to selecect multiple files.params.assembly-- set to "GRCh37" or "GRCh38".params.vep_cache-- full path to your localvep_cachedirectory.params.vep_flags-- flags you want to pass to VEP.params.loftee_flags-- comma-separated list of additional LoFtee flags (with leading comma). Flagsloftee_path,gerp_bigwig,human_ancestor_fa, andconservation_fileare set automatically based on the selectedassembly.enable_summary-- set totrueif you want to generate HTML summary files.process.container-- full path to theSingularityimage file (see step 1.1.).executor.$slurm.queueSize-- maximal number of SLURM jobs to submit at once.
-
Run pipeline:
module load nextflow module load apptainer nextflow run Annotation.nf -w ~/scratch/work_directoryImportant: when working on Compute Canada HPC, set working directory to ~/scratch/<new directory name>. This will speed up IO and also save space on your
projectpartition. After the execution, if there were no errors and you are happy with the results, you can remove this working directory.
In this section, we will explain how to integrate custom VCF files, such as gnomAD v2, into your VEP command line using this pipeline.
- Download the custom VCF into the cache. As an exemple, we downloaded the gnomAD v2 VCF file which is located at:
/path/to/vep_cache/custom_vcf/gnomad.exomes.r2.1.1.sites.liftover_grch38.PASS.noVEP.vcf.gz
- To integrate the custom VCF into your VEP command, add the relevant flags to the
nextflow.configfile under the--customflag. Below is an example configuration:
vep_flags = "--sift b --polyphen b --ccds --uniprot --hgvs --symbol --numbers --domains --regulatory --canonical --protein --biotype --af --af_1kg --af_gnomade --af_gnomadg --pubmed --shift_hgvs 0 --allele_number --buffer_size 10000 --custom /opt/vep/.vep/custom_vcf/gnomad.exomes.r2.1.1.sites.liftover_grch38.PASS.noVEP.vcf.gz,gnomad_exomes,vcf,exact,0,AN_nfe,AC_nfe,non_cancer_AN_nfe,non_cancer_AC_nfegnomad.exomes.r2.1.1.sites.liftover_grch38.PASS.noVEP.vcf.gz,gnomad_exomes,vcf,exact,0,AN_nfe,AC_nfe,non_cancer_AN_nfe,non_cancer_AC_nfe"
In this case, the custom flag integrates the gnomAD v2 VCF with the following parameters:
path: Path to the custom VCF file.
identifier: A name for the custom annotation (e.g., gnomad_exomes).
type: The file type (e.g., vcf).
match_type: How to match the data (e.g., exact).
cols: Columns to include from the VCF (e.g., 0,AN_nfe,AC_nfe,non_cancer_AN_nfe,non_cancer_AC_nfe).
- Run pipeline. Once you have updated your nextflow.config, run your VEP command as usual. The custom VCF annotations will be included in the output.
-
You may not be able to execute
nextflowdirectly from the Compute Canada login nodes due to the 8Gb memory limit per user. One alternative is to start interactive slurm job and submit all commands from it e.g.:salloc --time=2:00:00 --ntasks=1 --mem-per-cpu=16GOr submit a batch job with nextflow command e.g.:
module load nextflow module load apptainer sbatch --time=2:00:00 --ntasks=1 --mem-per-cpu=16G --wrap="nextflow run Annotation.nf -w ~/scratch/work_directory"Make sure you specify enough time. VEP annotation is typically fast, but total
nextflowexecution time will depend on how busy the SLURM queue is. -
Sometimes
nextflowwill crash with errorFailed to submit process to grid scheduler for execution. Most probably the SLURM queue was too busy and thus slow to respond. Your results were not lost, just resumenextflowexecution with the following command andnextflowwill continue from where it finished:nextflow run Annotate.nf -w ~/scratch/work_directory -resume -
If
nextflowcrashes with errorlibnet.so: failed to map segment from shared object, then try to increase the amount of memory in yoursallocorsbatchjob.