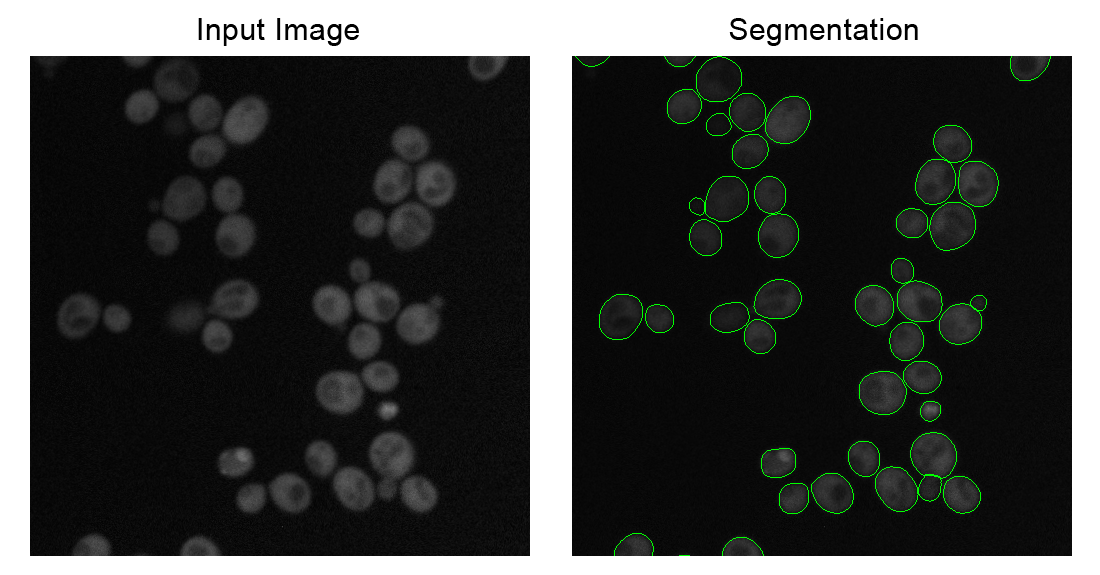
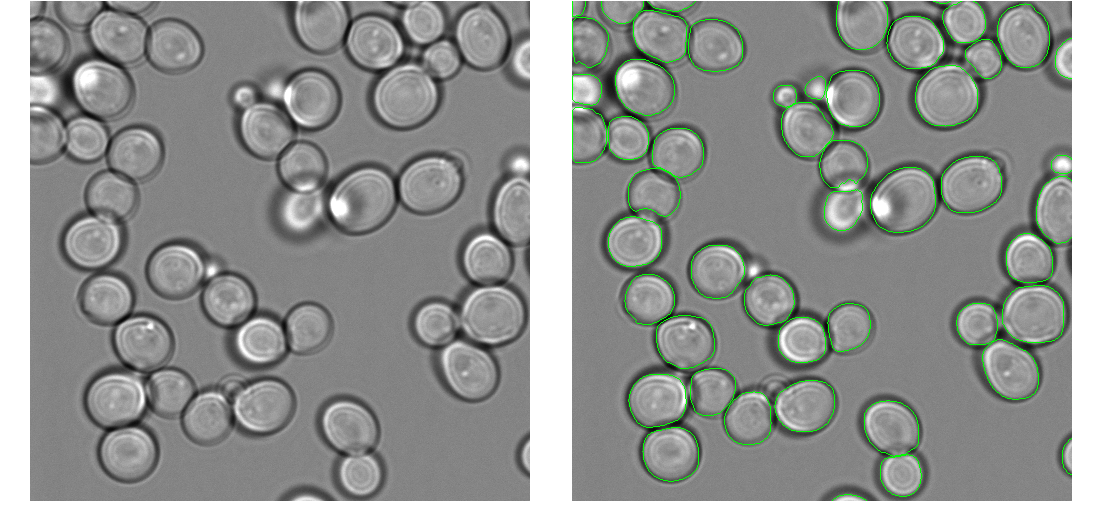
Code to segment yeast cells using a pre-trained mask-rcnn model. We've tested this with yeast cells imaged in fluorescent images and brightfield images, and gotten good results with both modalities. This code implements an user-friendly script that hides all of the messy implementation details and parameters. Simply put all of your images to be segmented into the same directory, and then plug and go.
(Adapted from the code of Deep Retina from the Kaggle 2018 Data Science Bowl, all credit for training models and wrangling the Mask-RCNN code goes to them: https://github.com/Lopezurrutia/DSB_2018)
We've received a few messages that this code breaks with the latest versions of Keras/Tensorflow, since their imports are in different places now. We'll eventually get around to releasing an updated version that works with more recent versions (when I finally get my thesis done and submitted, probably!), but for now, downgrading your packages to the minimal versions specified in requirements.txt will fix these import errors.
For this model to work, you will need the pre-trained weights for the neural network, which are too large to share on GitHub. You can grab them using following commands.
cd yeast_segmentation
wget https://zenodo.org/record/3598690/files/weights.zip # 226MB
unzip weights.zip
Once downloaded, simply unzip, so that there is a sub-directory called "weights" containing a file called "deepretina_final.h5" in the same directory of the segmentation.py and opts.py scripts.
conda create --name yeast_segmentation python=3.6 ## create an environment
conda activate yeast_segmentation ## activate the environment
cd /path/to/yeast_segmentation ## change directory to yeast_segmentation
pip install -r requirements.txt ## install requirements
Run segmentation without having to organise the images in a particular way (developments in this fork).
conda activate yeast_segmentation ## activate the environment
cd /path/to/yeast_segmentation ## change directory to yeast_segmentation
python run.py batch --help
usage: run.py batch [-h] [-f FUN_RENAME_INPUT] [-r] [--scale-factor SCALE_FACTOR] [--save-preprocessed] [--save-compressed] [--save-masks] [-v] [-u] [-y YEASTSPOTTER_SRCD] [-t]
input_directory output_directory brightps
Run yeastspotter.
positional arguments:
input_directory - path to the input directory.
output_directory - path to the output directory.
brightps - list of paths of the raw images to segment.
optional arguments:
-h, --help show this help message and exit
-
-r, --rescale True
--scale-factor SCALE_FACTOR
2
--save-preprocessed False
--save-compressed False
--save-masks True
-v, --verbose True
-u, --use-opts False
-y YEASTSPOTTER_SRCD, --yeastspotter-srcd path to the cloned repository.
-
-t, --test False
## set yeast_segmentation environment
import sys
sys.path.append('/path/to/yeast_segmentation')
from yeast_segmentation.run import batch
brightp2copyfromto=batch(
yeastspotter_srcd='/path/to/yeast_segmentation',
input_directory='inputs/',
output_directory='outputs/',
brightps=['image.{tiff|npy}'],
)
- Put all of the images you want to segment in one directory (this will be the input directory).
- Modify the opts.py file to point to the input directory, as well as the output directory to write the segmentations to (this directory should be empty). You can change various settings here, including what immediate files to write, and whether to rescale the images during segmentation to speed up the process.
- Run "python segmentation.py", and the script should handle everything from preprocessing to feeding into the neural network to post-processing. You're done!
- The output will be an integer-labeled image. Background is assigned a value of 0, and each individual cell is assigned an integer in incremental order (e.g. 1, 2, 3...) Stay tuned for some scripts to upload these files into ImageJ, Matlab, and Python for subsequent analysis soon.
These examples were produced from the same model, with the same parameters. There is no need to manually specify any parameters when switching between modalities or images from different microscopes.