Rui Xiao May 3, 2019
- Read the raw count-table and sample annotation files
- Preprocess the input data
- Principal Component Analysis
- Perform pairwise comparisons
- Fit generalized linear model
if (!requireNamespace("BiocManager",quietly=T)) install.packages("BiocManager")
library(BiocManager)
c1=read.table("counts_table_raw.txt",sep="\t",header=T,stringsAsFactors=F,row.names=1)
c2=read.table("counts_table_raw set 2.txt",sep="\t",header=T,stringsAsFactors=F,row.names=1)
c=cbind(c1,c2) # knowing that the row names are matching perfectly
rm(c1,c2) # free memory if no longer needed
dim(c)## [1] 23420 24
head(c) # count table overview## JK001 JK004 JK005 JK011 JK012 JK014 JK017 JK018 JK021 JK022 JK023
## Xkr4 2 0 0 0 5 2 1 1 0 0 0
## Rp1 76 5 89 81 34 141 100 43 25 248 173
## Sox17 1119 912 1169 417 1401 433 554 201 483 355 113
## Mrpl15 678 539 586 465 765 395 428 783 612 493 515
## Lypla1 1434 969 1378 963 1746 856 719 1280 1200 1190 1173
## Tcea1 877 866 785 688 938 593 685 739 692 772 817
## JK028 JK031 JK036 JK039 JK040 JK215 JK217 JK218 JK219 JK228 JK229
## Xkr4 5 0 0 0 0 15 7 5 10 9 11
## Rp1 18 106 131 2 4 154 305 56 122 20 31
## Sox17 427 440 612 327 616 2046 2288 1202 1614 675 663
## Mrpl15 1110 489 428 1160 998 926 1180 522 943 1101 968
## Lypla1 1788 1110 931 1270 1145 2791 3623 1494 2783 2574 2407
## Tcea1 1196 813 654 1051 857 1773 2519 1132 1853 1705 1694
## JK230 JK231
## Xkr4 5 15
## Rp1 162 99
## Sox17 1514 982
## Mrpl15 1000 717
## Lypla1 2974 2216
## Tcea1 1841 1354
a=read.csv("SampleAnnotation2Batches.csv",header=T,stringsAsFactors=F,row.names=1)
a$SingleFactor=paste(a$Batch,a$MMP13,a$SMOKE,a$FLU,sep="_") # merge factors into one for some further analysis
a$Batch=factor(a$Batch,levels=c(1,2))
a$MMP13=factor(a$MMP13,levels=c("WT","KO"))
a$FLU=factor(a$FLU,levels=c("PBS","FLU"))
a$SMOKE=factor(a$SMOKE,levels=c("RA","SM"))
head(a) # sample annotation overview## Batch MMP13 SMOKE FLU DOB SAC_DATE SingleFactor
## JK001 1 KO RA PBS 2/27/2017 6/26/2017 1_KO_RA_PBS
## JK004 1 KO RA PBS 2/27/2017 6/26/2017 1_KO_RA_PBS
## JK005 1 KO RA PBS 2/27/2017 6/26/2017 1_KO_RA_PBS
## JK011 1 WT SM PBS 2/27/2017 6/29/2017 1_WT_SM_PBS
## JK012 1 KO RA FLU 2/27/2017 6/30/2017 1_KO_RA_FLU
## JK014 1 WT SM PBS 2/27/2017 6/29/2017 1_WT_SM_PBS
if (!require("edgeR",quietly=T)) BiocManager::install("edgeR")
library(edgeR)
y=DGEList(counts=c,samples=a,group=a$SingleFactor)
keep <- rowSums(cpm(y)>1) >= 2
y <- y[keep, , keep.lib.sizes=FALSE]
y <- calcNormFactors(y)
head(y$samples[,2:3])## lib.size norm.factors
## JK001 20100075 0.9927551
## JK004 13444148 0.9744933
## JK005 19755033 1.0433833
## JK011 17655062 0.9687881
## JK012 21767921 1.0802167
## JK014 14472144 1.0178222
logcpm <- cpm(y, prior.count=2, log=TRUE)
logfile="logcpm.csv"
if (!file.exists(logfile)) write.csv(logcpm,logfile)
ifSig=function(logFC,P){ # P = PValue or FDR
return(abs(logFC)>=1&P<0.05)
# return(abs(logFC)>=0.5849&P<0.1)
}
# setwd("2018 results") # allow results to be written into the subfolder
if (!require("ggplot2")) install.packages("ggplot2")
library(ggplot2)if (!require("FactoMineR",quietly=T)) install.packages("FactoMineR")
library(FactoMineR)
al=cbind(a,t(logcpm))
colnames(al)[1:10] # annotaion goes from 1st to 7th column## [1] "Batch" "MMP13" "SMOKE" "FLU"
## [5] "DOB" "SAC_DATE" "SingleFactor" "Rp1"
## [9] "Sox17" "Mrpl15"
# multiple factors
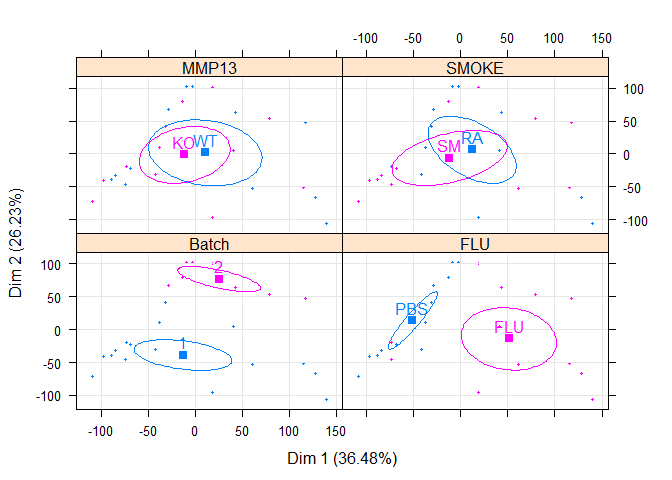
m.pca=PCA(al[,-c(5:7)],scale.unit=T,ncp=5,quali.sup=1:4,graph=F)
plotellipses(m.pca)# single factor
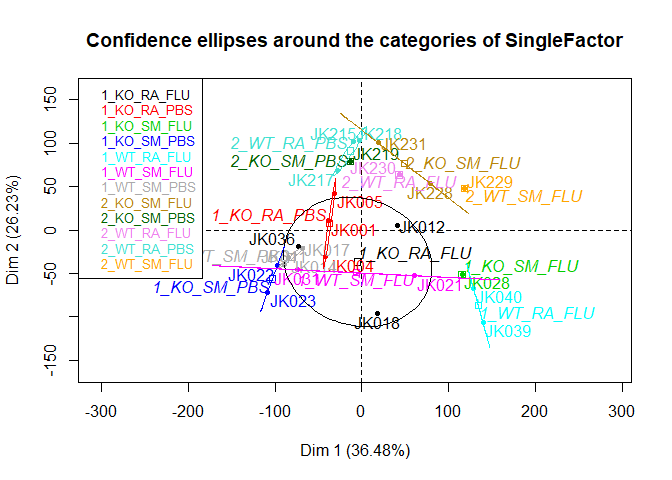
s.pca=PCA(al[,-c(1:6)],scale.unit=T,ncp=5,quali.sup=1,graph=F)
# plot.PCA(s.pca,axes=c(1,2),choix="ind",habillage = 1)
plotellipses(s.pca)if (!require("dplyr",quietly=T)) install.packages("dplyr")
library(dplyr) # %>%, filter, ...
if (!require("tibble",quietly=T)) install.packages("tibble")
library(tibble) # rownames_to_column('gene') column_to_rownames('gene')
if (!require("ggrepel")) install.packages("ggrepel")
library(ggrepel) # dodge labels for ggplot2
y1 <- estimateDisp(y,model.matrix(~SingleFactor,data=a))
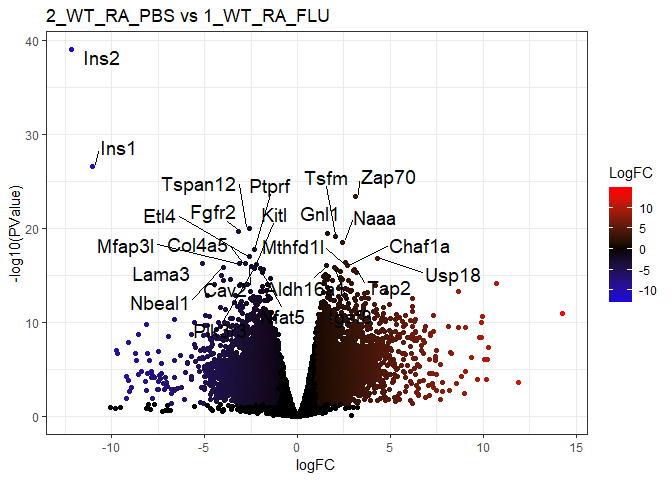
pairs <- rbind( # c("A","B") = B - A
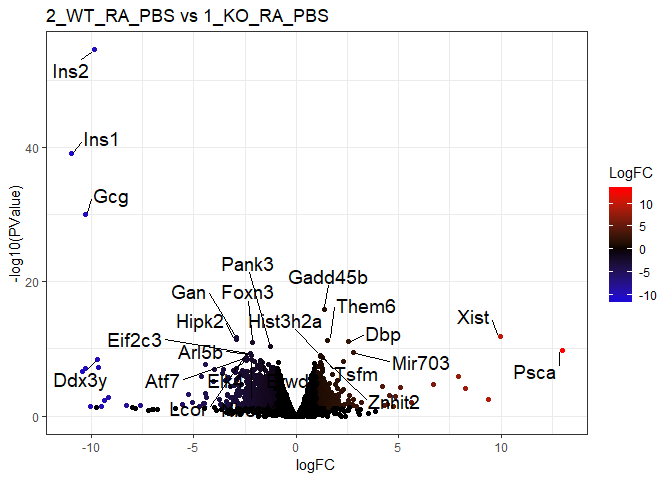
c("2_WT_RA_PBS","1_KO_RA_PBS"), # MMP13
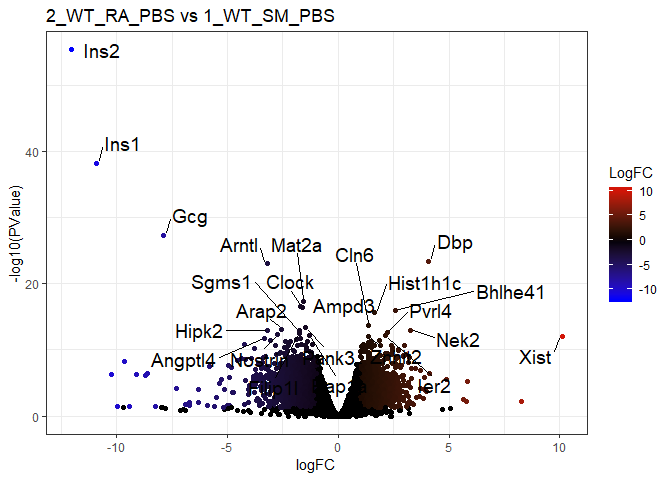
c("2_WT_RA_PBS","1_WT_SM_PBS"), # Smoke
c("2_WT_RA_PBS","1_WT_RA_FLU") # Flu
)
for (r in 1:nrow(pairs)) {
title=paste(pairs[r,],collapse=" vs ")
tt=topTags(exactTest(y1, pairs[r,]),n=9999999,sort.by="none") # select all with original order
# write.csv(tt,paste0("Pair_",title,".csv")) # save to local file
tt$table$LogFC=with(tt$table,ifelse(ifSig(logFC,PValue),logFC,0))
p = ggplot(tt$table,aes(logFC,-log10(PValue)))+
geom_point(aes(col=LogFC))+
scale_colour_gradient2(low="blue",mid="black",high="red")+
labs(title=title)+theme_bw()
# p
tt_filter_sort_top=(tt$table %>%
rownames_to_column('gene') %>%
filter(LogFC!=0) %>%
arrange(FDR) %>%
column_to_rownames('gene'))[1:24,] # top_n() may need to ungroup
print(
p+geom_text_repel(
data=tt_filter_sort_top,
aes(label=row.names(tt_filter_sort_top)),size=5,
box.padding = unit(0.35, "lines"),point.padding = unit(0.3, "lines")
)
)
}# design=model.matrix(~MMP13+SMOKE+FLU,data=a)
# design=model.matrix(~MMP13*SMOKE*FLU,data=a)
design=model.matrix(~MMP13*SMOKE*FLU+Batch,data=a)
# design=model.matrix(~MMP13*SMOKE*FLU*Batch,data=a); design=design[,-c(13:16)]
y2 <- estimateDisp(y,design)
fit <- glmQLFit(y2, design)
allList=function(cf){ # select complete unordered test result regarding each coefficient
dt=as.data.frame(topTags(glmQLFTest(fit,coef=cf),n=999999,sort.by="none"))
colnames(dt)=paste0(colnames(design)[cf],"_",colnames(dt))
return(dt)
}
# write.csv(cbind(allList(2),allList(3),allList(4),allList(5),allList(6),allList(7),allList(8)),"GLM_All.csv") # combine and write to csv file# functions that select stats on each factor
filterCount=function(cf){
cat(paste0(colnames(design)[cf]," Counts:\n"))
return(nrow(subset(as.data.frame(topTags(glmQLFTest(fit, coef=cf),n=999999)),ifSig(logFC,FDR))))
}
filterTop=function(cf,top=24){
cat(paste0(colnames(design)[cf]," top",top," :\n"))
return(topTags(glmQLFTest(fit, coef=cf),n=top))
}
filterName=function(cf){
cat(paste0(colnames(design)[cf]," Name:\n"))
return(row.names(subset(as.data.frame(topTags(glmQLFTest(fit, coef=cf),n=999999)),ifSig(logFC,FDR))))
}
filterWrite=function(cf){
cat(paste0(colnames(design)[cf]," written to file:\n"))
write.csv(subset(as.data.frame(topTags(glmQLFTest(fit, coef=cf),n=999999)),ifSig(logFC,FDR)),
paste0("",gsub(":","+",colnames(design)[cf]),".csv"))
}
for (i in 2:ncol(design)){ # uncomment following lines to enable the outputs
cat(filterCount(i),end="\n")
# print(filterTop(i))
# print(filterName(i))
# filterWrite(i)
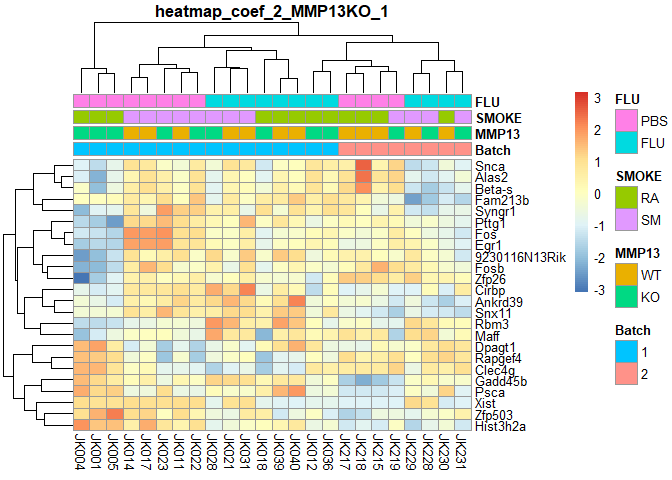
}## MMP13KO Counts:
## 1
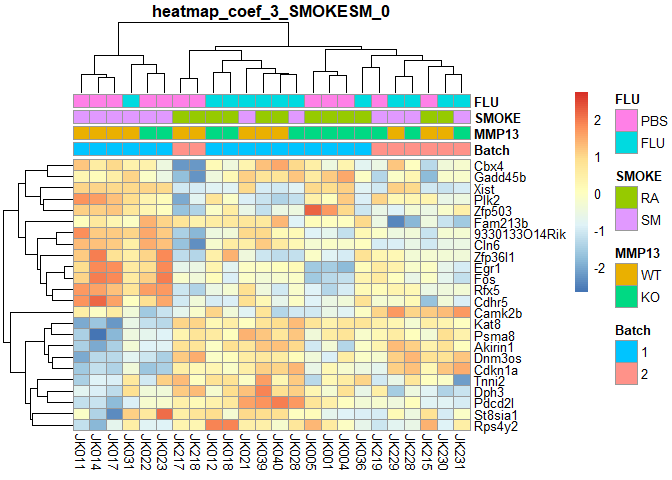
## SMOKESM Counts:
## 0
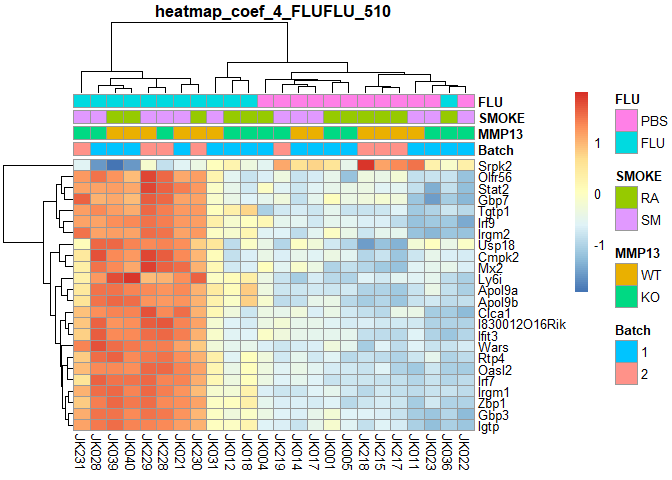
## FLUFLU Counts:
## 510
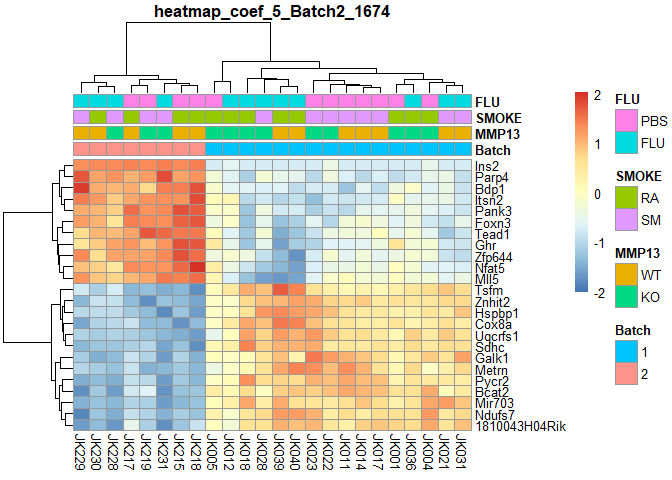
## Batch2 Counts:
## 1674
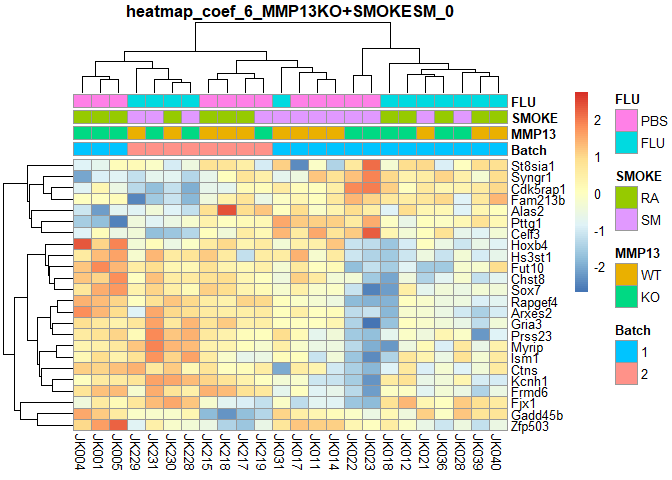
## MMP13KO:SMOKESM Counts:
## 0
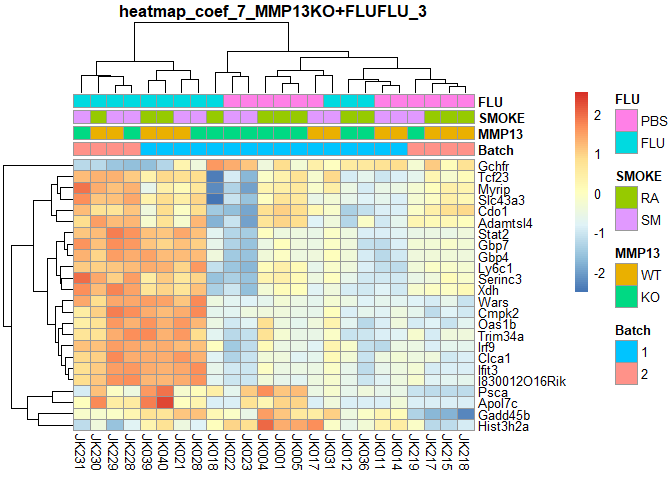
## MMP13KO:FLUFLU Counts:
## 3
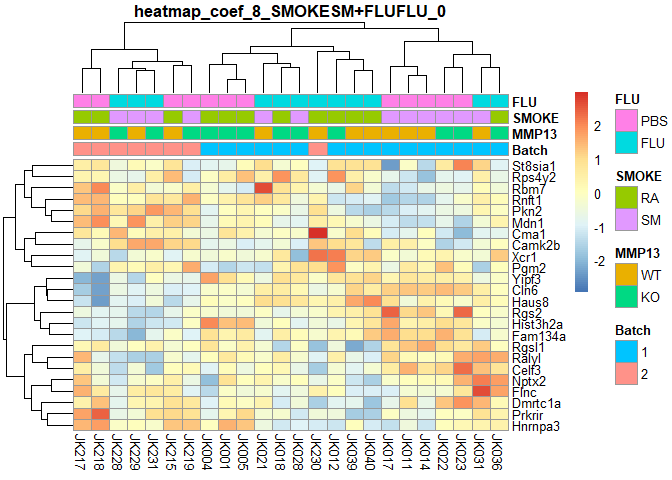
## SMOKESM:FLUFLU Counts:
## 0
## MMP13KO:SMOKESM:FLUFLU Counts:
## 2
if (!require("pheatmap",quietly=T)) install.packages("pheatmap")
library(pheatmap)
order=a[order(a$Batch,a$MMP13,a$SMOKE,a$FLU),] # ordered annotation
logcpm=logcpm[,row.names(order)] # ordered logcpm
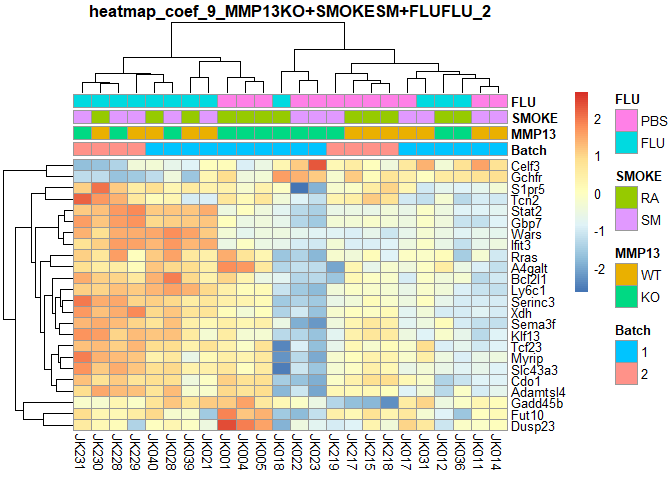
plotPheatmap=function(cf,clus_col=T,pdf=T){
d=as.data.frame(logcpm[row.names(filterTop(cf)),])
outfile=gsub(":","+",paste("heatmap","coef",i,colnames(design)[i],filterCount(cf),sep="_"))
if (pdf) pdf(paste0(outfile,".pdf"),width=9,height=6)
pheatmap(d,scale="row",annotation_col=order[,1:4],cluster_cols=clus_col,main=outfile)
if (pdf) dev.off()
}
for (i in 2:ncol(design)){
plotPheatmap(i,pdf=F)
}## MMP13KO top24 :
## MMP13KO Counts:
## SMOKESM top24 :
## SMOKESM Counts:
## FLUFLU top24 :
## FLUFLU Counts:
## Batch2 top24 :
## Batch2 Counts:
## MMP13KO:SMOKESM top24 :
## MMP13KO:SMOKESM Counts:
## MMP13KO:FLUFLU top24 :
## MMP13KO:FLUFLU Counts:
## SMOKESM:FLUFLU top24 :
## SMOKESM:FLUFLU Counts:
## MMP13KO:SMOKESM:FLUFLU top24 :
## MMP13KO:SMOKESM:FLUFLU Counts:
if (!require("systemPipeR",quietly=T)) BiocManager::install("systemPipeR")
library(systemPipeR)
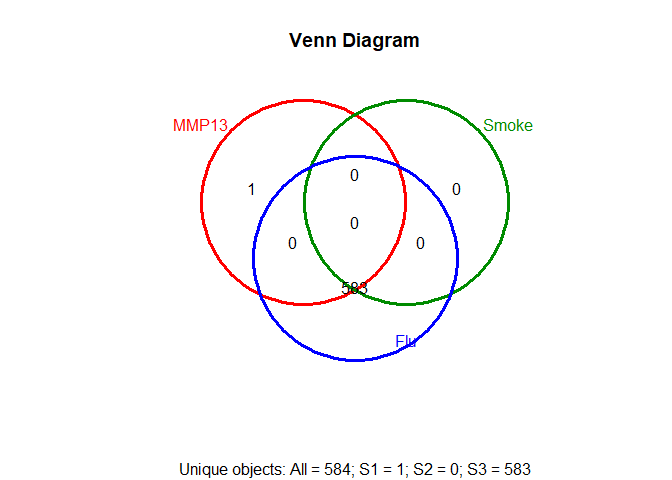
setlist=list(MMP13=row.names(topTags(glmQLFTest(fit, coef=2),p.value=0.05,n=999999)),
Smoke=row.names(topTags(glmQLFTest(fit, coef=3),p.value=0.05,n=999999)),
Flu=row.names(topTags(glmQLFTest(fit, coef=4),p.value=0.05,n=999999)))
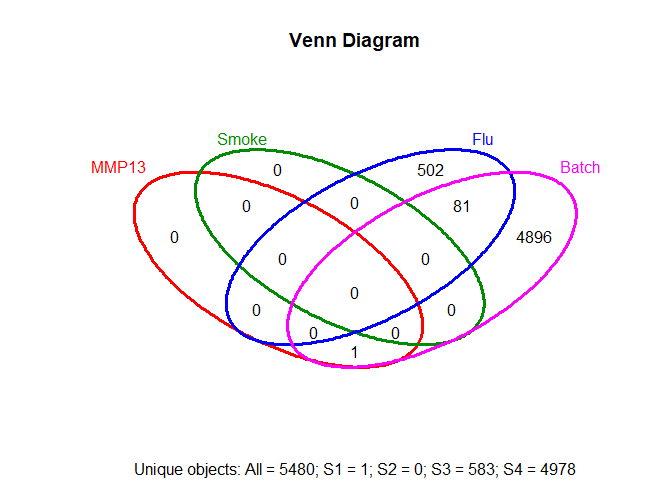
vennPlot(overLapper(setlist,type="vennsets"))setlist=list(MMP13=row.names(topTags(glmQLFTest(fit, coef=2),p.value=0.05,n=999999)),
Smoke=row.names(topTags(glmQLFTest(fit, coef=3),p.value=0.05,n=999999)),
Flu=row.names(topTags(glmQLFTest(fit, coef=4),p.value=0.05,n=999999)),
Batch=row.names(topTags(glmQLFTest(fit, coef=5),p.value=0.05,n=999999)))
vennPlot(overLapper(setlist,type="vennsets"))Data credits: Jeanine D’Armiento, Monica Goldklang, Kyle Stearns; Columbia University Medical Center