Welcome to this R shiny app!
Update (2024 Jan 31)
Bioconductor package STAN local installation is avaliable with 'STAN.zip' (genoSTAN 2.24.0):
install.packages("YOUR_PATH/STAN", repo = NULL, type = "source")
Transcription unit (TU) annotation tool for nascent RNA-Seq analysis.
Usage:
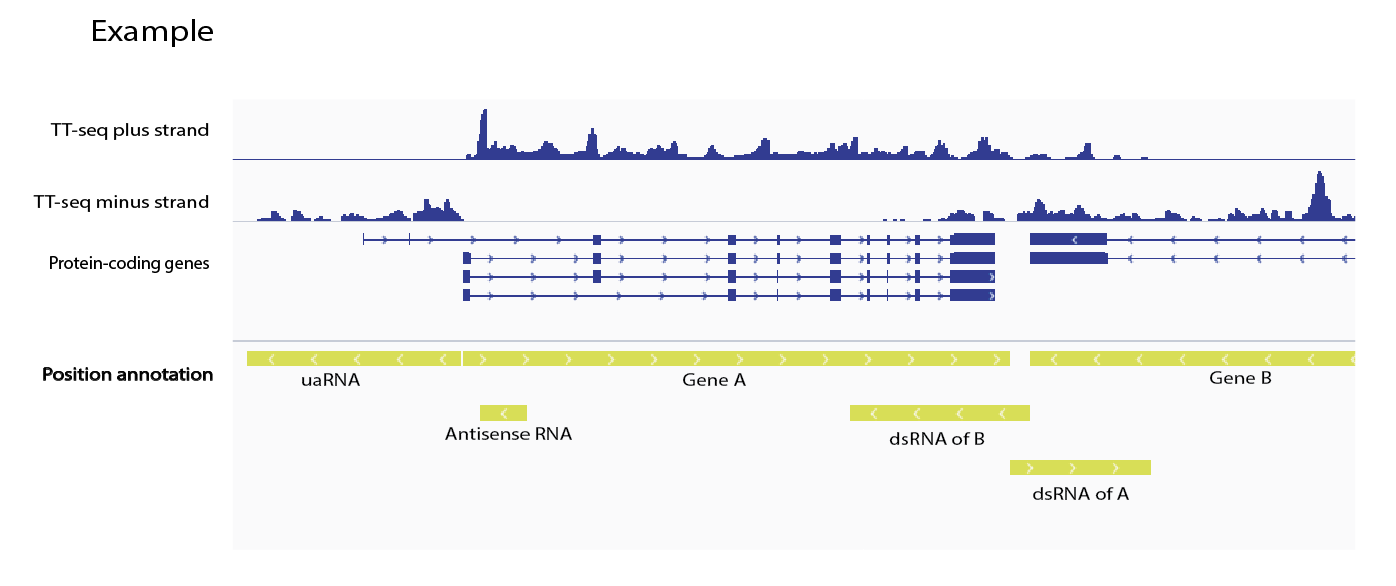
- transcribed genomic regions (e.g. non-coding intergenic RNAs)
- multiple transcriptomes comparison (e.g. GRO-seq vs PRO-seq)
- alternative to HOMER's GRO-seq analysis, with an optional mRNA TU merging step (recommended).
Inputs:
- gene reference (GENCODE or ensembl)
- bam files (allow multiple files from replicates)
Output:
- gff3 files of TU intervals (in a zipped folder)
- with columns: expression, gene id, transript id, exon overlap, gene type, relative location to gene.
Non-coding TUs will be named according to their relative locations to the neighbored genes.
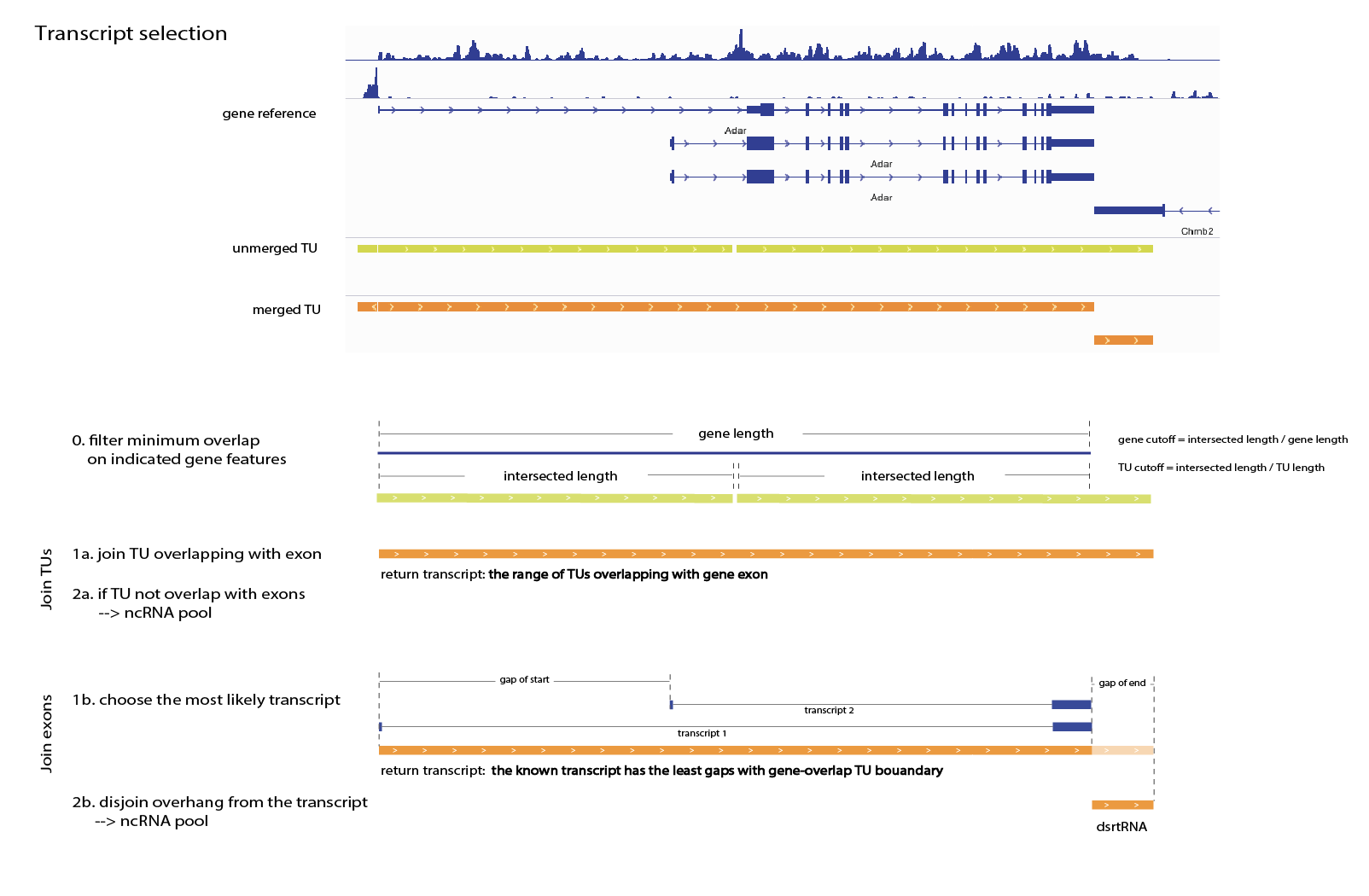
Scattered TU intervals overlapping with gene references will be joined with a selected method:
- by TU (3' termination region will often be included)
- by exon (only keeps TU intervals within gene boundary, for accurate expression level estimation, but also produces trimmed flanking TUs)