preqclr is a software tool that reports on quality for long read sequencing data without the use of a reference genome. With the emergence of new long read sequencing technology such as Pacbio Single Molecule, Real-Time (SMRT) Sequencing technology and Oxford Nanopore Technologies (ONT), there is a need for a method that assesses sequencing quality prior to analyses. This tool enables users to visualize metrics of quality.
There are two components to preqclr:
1. calculate
2. report
preqclr generates a PDF report with the following plots:
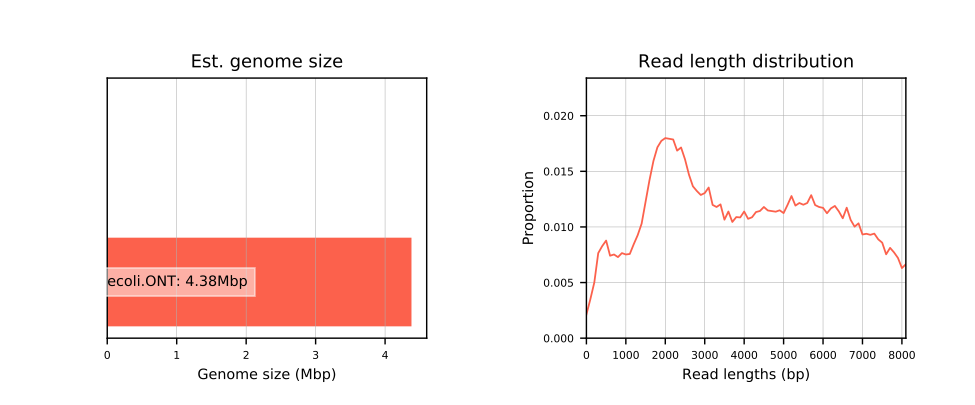
- Estimated genome size
- Read length distribution
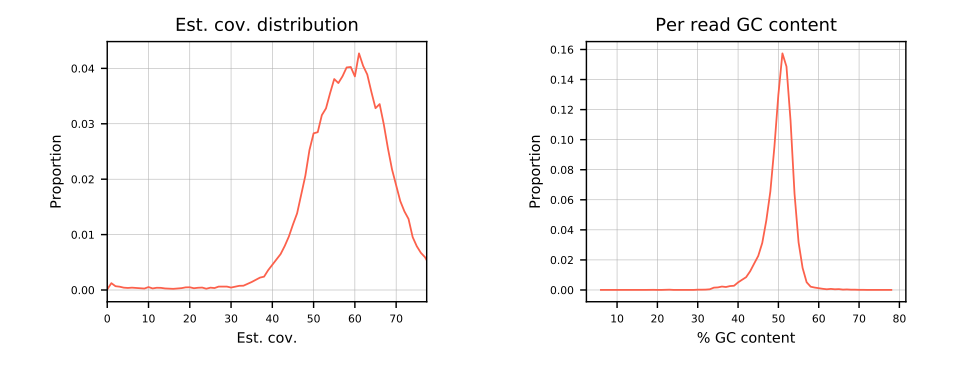
- Estimated coverage distribution
- Per read GC content distribution
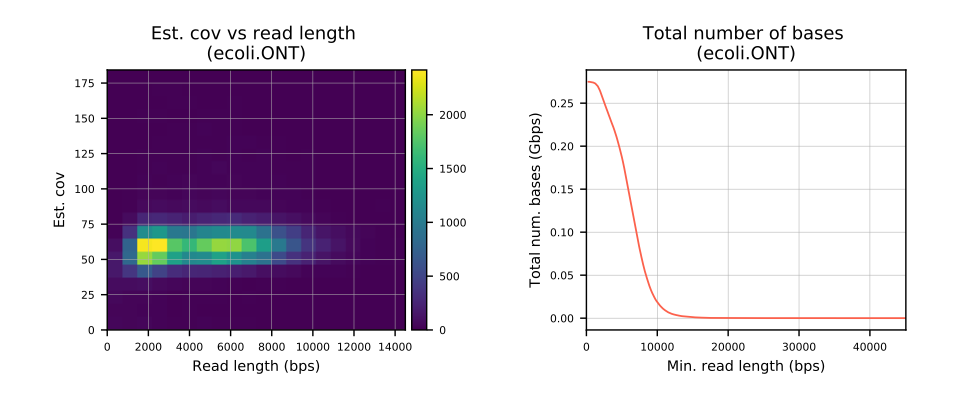
- Estimated coverage vs read length
- Total number of bases as a function of minimum read length
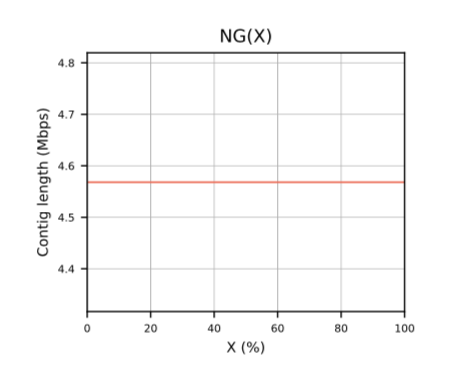
- NG(X)
To create the files needed:
- minimap2 (for overlap detection)
- miniasm (optional for NGX plots)
For the calculation step:
- C++ compiler with C++11 support
For the report generation step:
- Python2.7.11
- matplotlib
- BioPython
- setuptools (to download report script dependencies)
To install from source:
git clone --recursive https://github.com/simpsonlab/preqclr.git
cd preqc-lr
make
# download report script dependencies
# create virtual environment
virtualenv preqclr-venv
source preqclr-venv/bin/activate
python setup.py install# STEP 0: overlap detection and contig lengths
minimap2 -x ava-ont reads.fq reads.fq > overlaps.paf
miniasm -f reads.fq overlaps.paf > layout.gfa
# STEP 1: calculate data for plots
./preqclr -r reads.fq \
--paf overlaps.paf \
--gfa layout.gfa \
-n ecoli_sample.pacbio \
--verbose
# STEP 2: create a PDF report
python preqclr-report.py -i ecoli_sample.pacbio.preqclr --verbose - When using minimaps, we recommend using the settings optimized for PacBio reads (
-x ava-pb) and ONT reads (-x ava-ont).
preqclr uses overlap and assembly information from minimap2 and miniasm, respectively. To parse the output PAF file, and efficiently read fasta files we used kseq and PAF parser in miniasm. I would like to thank Heng Li for developing these tools.