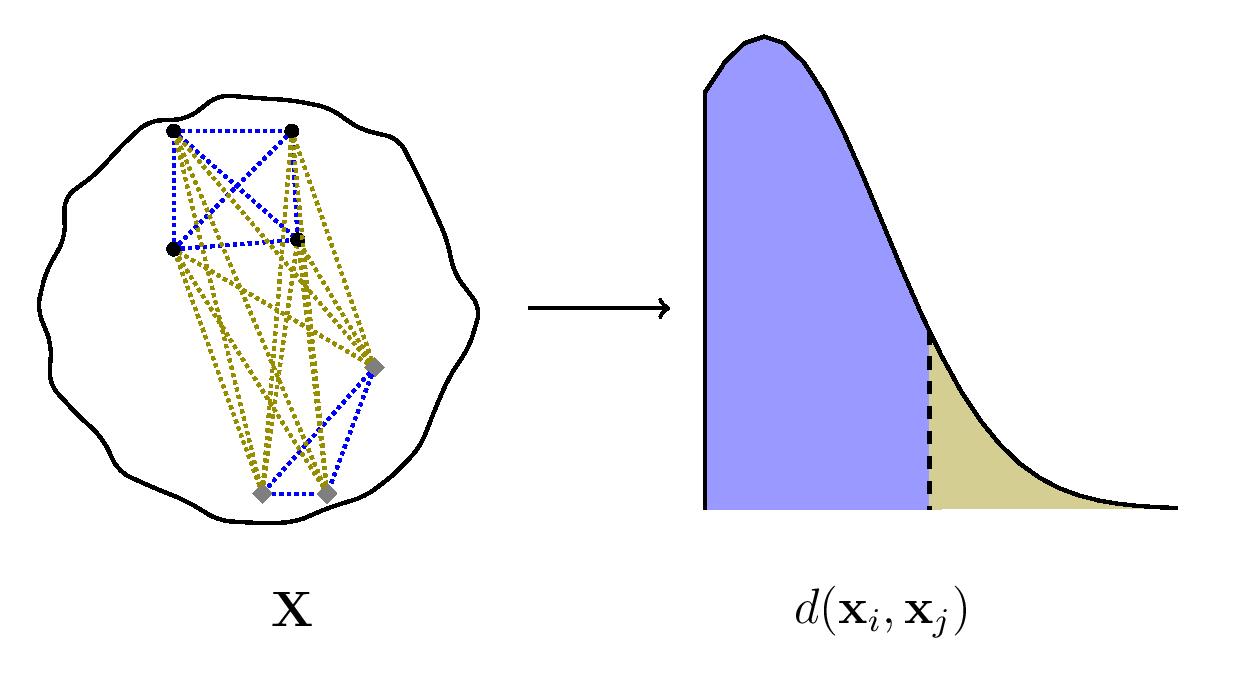
Given a cluster, dataset, or sample comprising multiple observations, the existence of subclusters, subgroups or stratification is accompanied by a wider spread of pairwise distances between the observations.
Welcome to the homepage of flinty, a method for assessing sample non-exchangeability, or heterogeneity, in multivariate datasets. Details of our method can be found in
Aw, Spence and Song (2024) "A simple and flexible test of sample exchangeability with applications to statistical genomics," Annals of Applied Statistics 18 (1): 858-881. DOI: 10.1214/23-AOAS1817. Preprint available at arXiv:2109.15261.
Please visit the package homepage for flintyPy here. Our homepage provides installation instructions, as well as examples for using our methods.
Please visit the package homepage for flintyR here. Our homepage provides installation instructions, as well as numerous tutorials that expand on analyses reported in our paper.
While our documentation above should help you install our packages, our software is also available in the software subdirectory.
Users unfamiliar with exchangeability can learn more about the topic here:
With the kind help of domain experts from biology and the social sciences, we also offer integrative tutorials on the following topics:
Please feel free to use these tutorials for research or teaching purposes. We welcome any feedback or suggestions that will help make our work more accessible.
The Supplementary Information file to our main text contains technical proofs that may be of interest to the reader. We have made it available in the manuscript subdirectory.