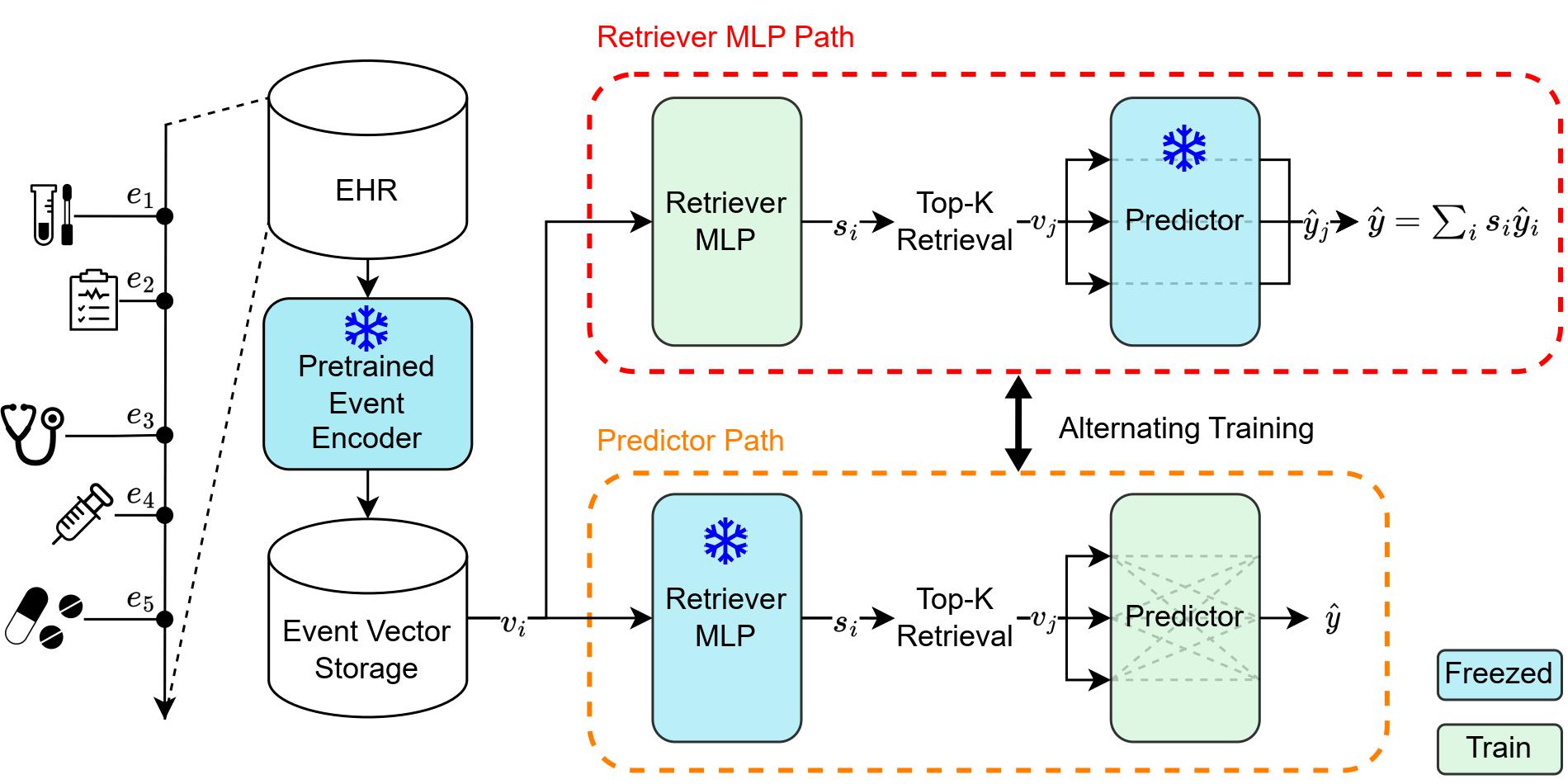
Official implementation for General-purpose Retrieval-Enhanced Medical Prediction Model using Near-Infinite History
Developing medical prediction models based on EHRs typically relies on expert opinion for feature selection and adjusting the observation window size. To address these issues, we propose Retrieval-Enhanced Medical prediction model (REMed), which can essentially evaluate an unlimited number of medical events, retrieve the relevant ones, and make predictions.
- For enhanced accessibility, we offer a simplified, standalone REMed model available in
standalone_remed.py. - This model takes a list of event vectors and their corresponding timestamps as input, and performs a binary classification.
- For multi-task or multi-class support, please refer to the original code.
- Dependencies:
pytorchandtransformers.
import torch
from standalone_remed import REMed
model = REMed(
pred_dim=512, # Model hidden dimension size (E)
n_heads=8, # Number of heads for Transformer Predictor
n_layers=2, # Number of layers for Transformer Predictor
dropout=0.2, # Dropout rate
max_retrieve_len=128, # Maximum number of retrieved events (k of Top-k)
pred_time=48, # Prediction time. Set to maximum of the input timestamp (h)
)
reprs = torch.randn(2, 1000, 512) # Batch of list of event vectors (B, L, E)
times = torch.randint(0, 48*60, (2, 1000)) # Batch of list of event times (B, L) (unit=Minute)
model(reprs, times) # Return probability between [0,1] (B, 1)Requirements
- For preprocessing:
python>=3.8, Java>=8
pip install numpy pandas tqdm treelib transformers pyspark- For training & test
export PATH=/usr/local/cuda/bin:$PATH
conda install pytorch==1.13.1 torchvision==0.14.1 torchaudio==0.13.1 pytorch-cuda=11.7 -c pytorch -c nvidia
conda install numpy pandas einops h5pickle tqdm scikit-learn -y
pip install performer_pytorch recurrent_memory_transformer_pytorch==0.2.2 transformers==4.30.1 accelerate==0.20.3
cd src/models/kernels/
python setup.py installData Preprocessing
- We use Integrated-EHR-Pipeline
- NOTE: This process requires high RAM. If you meet out-of-memory, please lower the
--num_threads
git clone https://github.com/Jwoo5/integrated-ehr-pipeline
git checkout snub# MIMIC-IV, 48h Prediction time
python main.py --ehr mimiciv --data {MIMIC-IV Path} --obs_size 48 --pred_size 48 --max_patient_token_len 2147483647 --max_event_size 2147483647 --use_more_tables --dest {DATA_PATH}/48h --num_threads 32 --readmission --diagnosis --min_event_size 0 --seed "2020, 2021, 2022, 2023, 2024" --use_ed
# MIMIC-IV, 24h Prediction time
python main.py --ehr mimiciv --data {MIMIC-IV Path} --obs_size 48 --pred_size 24 --max_patient_token_len 2147483647 --max_event_size 2147483647 --use_more_tables --dest {DATA_PATH}/24h --num_threads 32 --readmission --diagnosis --min_event_size 0 --seed "2020, 2021, 2022, 2023, 2024" --use_ed
# eICU, 48h Prediction time
python main.py --ehr eicu --data {eICU Path} --obs_size 48 --pred_size 48 --max_patient_token_len 2147483647 --max_event_size 2147483647 --use_more_tables --dest {DATA_PATH}/48h --num_threads 32 --readmission --diagnosis --min_event_size 0 --seed "2020, 2021, 2022, 2023, 2024"
# eICU, 24h Prediction time
python main.py --ehr eicu --data {eICU Path} --obs_size 48 --pred_size 24 --max_patient_token_len 2147483647 --max_event_size 2147483647 --use_more_tables --dest {DATA_PATH}/24h --num_threads 32 --readmission --diagnosis --min_event_size 0 --seed "2020, 2021, 2022, 2023, 2024"Pretrain Encoder & Caching
- We used NVIDIA RTX A6000 (48GB) for pretraining & Encoding
- If you meet CUDA OOM, please adjust the numbers in
src/main.py:270-271 - This requires large empty disk space (>200G)
accelerate launch --config_file config/single.json --num_processes 1 --gpu_ids {GPU_ID} main.py --src {SRC_DATA} --input {DATA_PATH} --save_dir {SAVE_PATH} --train_type short --time -99999 --pred_time {PRED_TIME} --wandb_project_name {PROJECT_NAME} --wandb_entity_name {ENTITY_NAME} --lr 5e-5 --random_sample --encode_events- As a result, you can get
{SRC_DATA}_encoded.h5at{SAVE_PATH}/{EXPERIMENT_NAME}.
Train REMed
- Note that the
{EXPERIMENT_NAME}refers to the name of the pre-training experiment. - If you want to run an experiment with infinite observation window, set time=-99999
- Otherwise, the time should be {PRED_TIME} - {OBS_SIZE} (e.g. pred time 48h, obs 12h -> time 36)
accelerate launch --config_file config/single.json --num_processes 1 --gpu_ids {GPU_ID} main.py --src {SRC_DATA} --input {DATA_PATH} --save_dir {SAVE_PATH} --train_type remed --time {TIME} --pred_time {PRED_TIME} --wandb_project_name {PROJECT_NAME} --wandb_entity_name {ENTITY_NAME} --lr 1e-5 --scorer --scorer_use_time --pretrained {EXPERIMENT_NAME} --no_pretrained_checkpoint@misc{kim2023generalpurpose,
title={General-Purpose Retrieval-Enhanced Medical Prediction Model Using Near-Infinite History},
author={Junu Kim and Chaeeun Shim and Bosco Seong Kyu Yang and Chami Im and Sung Yoon Lim and Han-Gil Jeong and Edward Choi},
year={2023},
eprint={2310.20204},
archivePrefix={arXiv},
primaryClass={cs.LG}
}