QUPS (pronounced "CUPS") is an abstract, lightweight, readable tool for prototyping pulse-echo ultrasound systems and algorithms. It provides a flexible, high-level representation of transducers, pulse sequences, imaging regions, and scattering media as well as hardware accelerated implementations of common signal processing functions for pulse-echo ultrasound systems. QUPS can interface with multiple other Ultrasound acquisition, simulation and processing tools including Verasonics, k-Wave, MUST, FieldII and USTB.
This package can readily be used to develop new transducer array designs by specifying element positions and orientations or develop new pulse sequence designs by specifying waveforms, element delays, and element weights (apodization). Simulating the received echoes (channel data) is supported for any valid UltrasoundSystem. Define custom properties or overload the built-in classes to create new types.
-
Flexible:
- 3D space implementation
- Transducers: Arbitrary transducer positions and orientations
- Sequences: Arbitrary transmit waveform, delays, and apodization
- Scans (Image Domain): Arbitrary pixel locations and beamforming apodization
-
Performant:
- Beamform a 1024 x 1024 image for 256 x 256 transmits/receives in < 2 seconds (RTX 3070)
- Hardware acceleration via CUDA (Nvidia) or OpenCL (AMD, Apple, Intel, Nvidia), or natively via the Parallel Computing Toolbox
- Memory efficient classes and methods such as separable beamforming delay and apodization ND-arrays minimize data storage and computational load
- Batch simulations locally via
parclusteror scale to a cluster with the MATLAB Parallel Server (optional) toolbox.
-
Modular:
- Transducer, pulse sequence, pulse waveform, scan region etc. each defined separately
- Optionally overload classes to customize behaviour
- Easily compare sequence to sequence or transducer to transducer
- Simulate with MUST, FieldII, or k-Wave without redefining most parameters
- Export or import data between USTB or Verasonics data structures
-
Intuitive:
- Native MATLAB semantics with argument validation and tab auto-completion
- Overloaded
plotandimagescfunctions for data visualization - Documentation via
helpanddoc
QUPS requires the Signal Processing Toolbox and the Parallel Computing Toolbox to be installed.
Starting in MATLAB R2023b+, QUPS and most of it's extension packages can be installed from within MATLAB via buildtool if you have setup git for MATLAB.
- Install qups
gitclone("https://github.com/thorstone25/qups.git");
cd qups;
- (optional) Install and patch extension packages and compile mex and CUDA binaries (failures can be safely ignored)
buildtool install patch compile -continueOnFailure
- (optional) Run tests (~10 min)
buildtool test
You can manually download and install each extension separately.
-
Download the desired extension packages into a folder adjacent to the "qups" folder e.g. if qups is located at
/path/to/my/qups, kWave should be downloaded to an adjacent folder/path/to/my/kWave. -
Create a MATLAB Project and add the root folder of the extension to the path e.g.
/path/to/my/kWave.
- Note: The "prj" file in USTB is a Toolbox file, not a Project file - you will still need to make a new Project.
-
Open the
Qups.prjproject and add each extension package as a reference. -
(optional) Apply patches to enable further parallel processing.
-
(optional) Run tests via the
runProjectTests()function in the build directory.
addpath build; runProjectTests('verbosity', 'Concise'),
All extensions to QUPS are optional, but must be installed separately from their respective sources.
| Extension | Description | Installation Paths | Citation |
|---|---|---|---|
| FieldII | point scatterer simulator | addpath /path/to/fieldII |
website |
| k-Wave | distributed medium simulator | addpath /path/to/kWave |
website |
| kWaveArray | k-Wave transducer extension | addpath /path/to/kWaveArray |
forum, paper |
| MUST | point scatterer simulator | addpath /path/to/MUST |
website |
| USTB | signal processing library and toolbox | addpath /path/to/USTB |
website |
| Matlab-OpenCL | hardware acceleration | (see README) | website (via MatCL) |
| CUDA | hardware acceleration | (see CUDA Support) |
- Start MATLAB R2020b or later and open the Project
openProject .
- (optional) Setup any available acceleration
setup parallel CUDA cache; % setup the environment with any available acceleration
- Create an ultrasound system and point scatterer to simulate
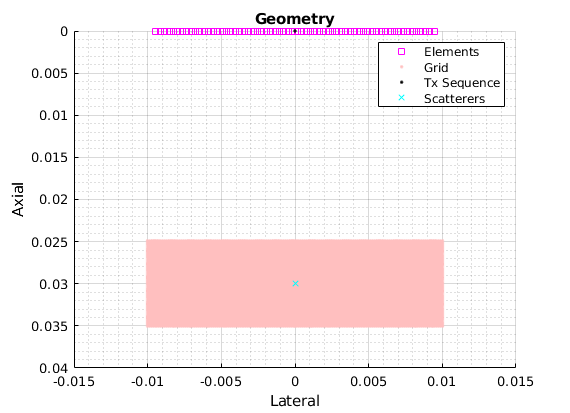
scat = Scatterers('pos', 1e-3*[0 0 30]'); % a single point scatterer at 20mm depth
xdc = TransducerArray.P4_2v(); % simulate a Verasonics L11-5v transducer
seq = Sequence('type', 'FSA', 'numPulse', xdc.numel); % full synthetic-aperture pulse sequence
scan = ScanCartesian('x', 1e-3*[-10, 10], 'z', 1e-3*[25 35]); % set the image boundaries - we'll set the resolution later
us = UltrasoundSystem('xdc', xdc, 'seq', seq, 'scan', scan, 'fs', 4*xdc.fc); % create a system description
[us.scan.dx, us.scan.dz] = deal(us.lambda / 4); % set the imaging resolution based on the wavelength
- Display the geometry
figure; plot(us); hold on; plot(scat, 'cx'); % plot the ultrasound system and the point scatterers
- Simulate channel data
chd = greens(us, scat); % create channel data using a shifted Green's function (CUDA/OpenCL-enabled)
% chd = calc_scat_multi(us, scat); % ... or with FieldII
% chd = kspaceFirstOrder(us, scat); % ... or with k-Wave (CUDA-enabled)
% chd = simus(us, scat); % ... or with MUST (CUDA-enabled)
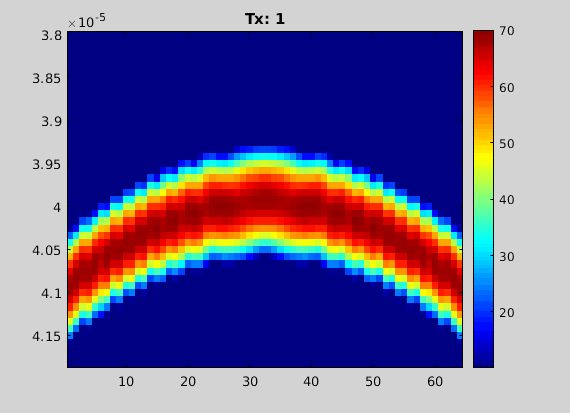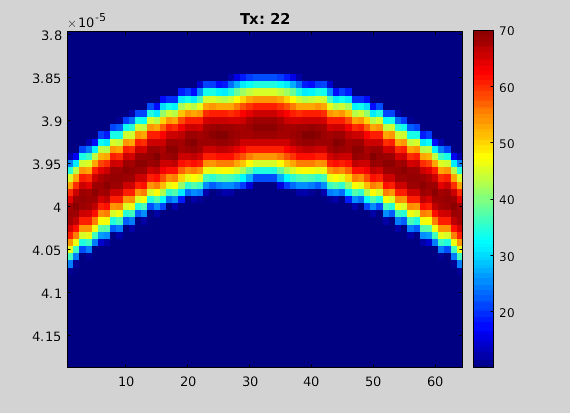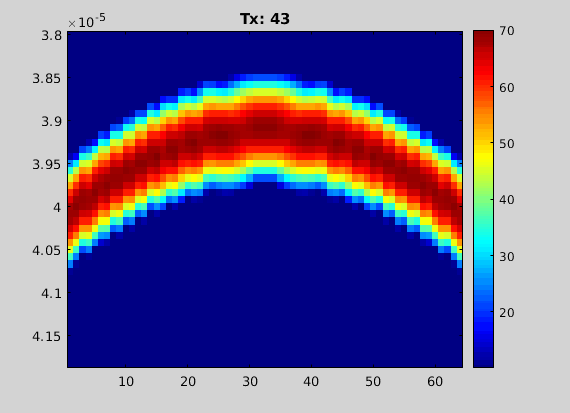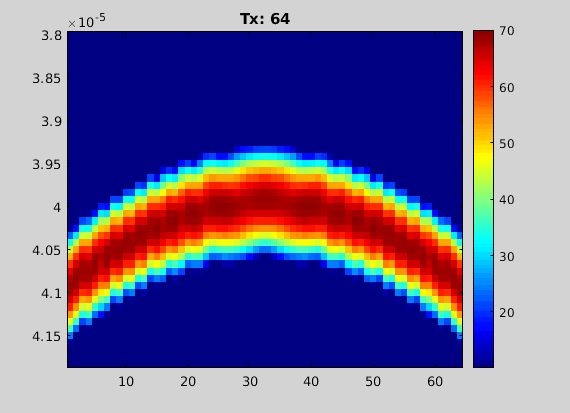
- Display the channel data
figure; imagesc(chd); dbr echo 60;
animate(chd.data, 'loop', false, 'title', "Tx: "+(1:chd.M));
- Beamform
b = DAS(us, hilbert(chd));
- Display the B-mode image
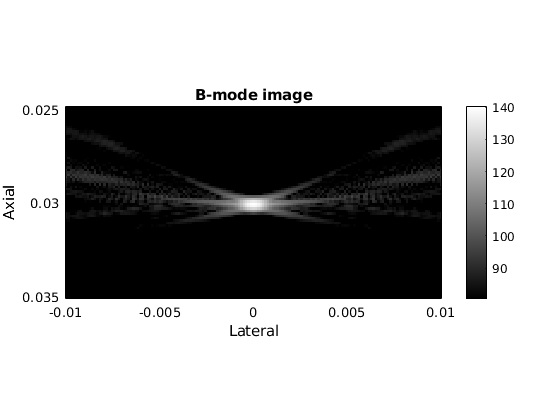
figure; imagesc(us.scan, b); dbr b-mode 60;
title('B-mode image');
QUPS is documented within MATLAB. To see all the available classes, use help ./src or doc ./src from within the QUPS folder. Use help or doc on any class or method with help classname or help classname.methodname e.g. help UltrasoundSystem.DAS.
For a walk through of going from defining a simulation to a beamformed image, see example.mlx (or example_.m).
See the examples folder for examples of specific applications.
For syntax examples for each class, see cheat_sheet.m.
For further documentation on customizing classes, see the class structure README.
If you have trouble, please submit an issue.
If you use this software, please cite this repository using the citation file or via the menu option in the "About" section of the github page.
If you use any of the extensions, please see their citation policies:
Some QUPS methods, including most simulation and beamforming methods, can be parallelized natively by specifying a parcluster or launching a parallel.ProcessPool or ideally a parallel.ThreadPool. However, restrictions apply.
Workers in a parallel.ThreadPool cannot call mex functions, use GUIs or user inputs, or perform any file operations (reading or writing) before R2024a. Workers in a parallel.ProcessPool or parcluster do not have these restrictions, but tend to be somewhat slower and require much more memory. All workers are subject to race conditions.
Removing race conditions and inaccesible functions in the extension packages will enable native parallelization. The patches described below are applied automatically with the "patch" task via buildtool. Otherwise, you will need to apply the patches manually to enable parallelization.
FieldII uses mex functions for all calls, which requires file I/O. This cannot be used with a parallel.ThreadPool, but can easily be used with a parallel.ProcessPool or parcluster.
k-Wave (with binaries)
To enable simulating multiple transmits simultaneously using k-Wave binaries, the temporary filename race condition in kspaceFirstOrder3DC.m must be remedied.
Edit kspaceFirstOrder3DC.m and look for an expression setting the temporary folder data_path = tempdir. Replace this with data_path = tempname; mkdir(data_path); to create a new temporary directory for each worker.
You may also want to delete this folder after the temporary files are deleted. Record a variable new_path = true; if a new directory was created, and place if new_path, rmdir(data_path); end at the end of the function. Otherwise, the temporary drive is cleared when the system reboots.
On Linux, the filesystem does not deallocate deleted temporary files until MATLAB is closed. This can lead to write erros if many large simulations are run in the same MATLAB session. To avoid this issue, within kspaceFirstOrder3DC.m, set the file size of the temporary input/output files to 0 bytes prior to deleting them, e.g.
if isunix % tolerate deferred deletion for parpools on linux
system("truncate -s 0 " + input_filename );
system("truncate -s 0 " + output_filename);
end
delete(input_filename );
delete(output_filename);
To enable the usage of a parallel.ThreadPool with the simus() method, the GUI and file I/O calls used in the AdMessage and MUSTStat functions must not be called from pfield.m and/or pfield3.m (see #2). It is safe to comment out the advertising and statistics functions.
OpenCL support is provided via Matlab-OpenCL, but is only tested on Linux. This package relies on MatCL, but the underlying OpenCL installation is platform and OS specific. The following packages and references may be helpful, but are not tested for compatability.
| Command | Description |
|---|---|
sudo apt install opencl-headers |
Compilation header files (req'd for all devices) |
sudo apt install pocl-opencl-icd |
Most CPU devices |
sudo apt install intel-opencl-icd |
Intel Graphics devices |
sudo apt install nvidia-driver-xxx |
Nvidia Graphics devices (included with the driver) |
sudo apt install ./amdgpu-install_x.x.x-x_all.deb |
AMD Discrete Graphics devices (see here or here) |
Starting in R2023a, CUDA support is provided by default within MATLAB via mexcuda.
Otherwise, for CUDA to work, nvcc must succesfully run from the MATLAB environment. If a Nvidia GPU is available and setup CUDA cache completes with no warnings, you're all set! If you have difficulty getting nvcc to work in MATLAB, you may need to figure out which environment paths are required for your CUDA installation. Running setup CUDA will attempt to do this for you, but may fail if you have a custom installation.
First, be sure you can run nvcc from a terminal or command-line interface per CUDA installation instructions. Then set the MW_NVCC_PATH environmental variable within MATLAB by running setenv('MW_NVCC_PATH', YOUR_NVCC_BIN_PATH); prior to running setup CUDA. You can run which nvcc within a terminal to locate the installation directory. For example, if which nvcc returns /opt/cuda/bin/nvcc, then run setenv('MW_NVCC_PATH', '/opt/cuda/bin');.
First, setup your system for CUDA per CUDA installation instructions. On Windows you must set the path for both CUDA and the correct MSVC compiler for C/C++. Start a PowerShell terminal within Visual Studio. Run echo %CUDA_PATH% to find the base CUDA_PATH and run echo %VCToolsInstallDir% to find the MSVC path. Then, in MATLAB, set these paths with setenv('MW_NVCC_PATH', YOUR_CUDA_BIN_PATH); setenv('VCToolsInstallDir', YOUR_MSVC_PATH);, where YOUR_CUDA_BIN_PATH is the path to the bin folder in the CUDA_PATH folder. Finally, run setup CUDA. From here the proper paths should be added.
Most functions will perform best with an active parallel.ThreadPool available. This can be started with setup parallel or parpool Threads. Alternatively, a parallel.ProcessPool can be started with parpool Processes (or parpool local on earlier MATLAB releases). Functions that allow specifying a parallel environment can avoid using the active parallel pool by using an argument of 0, or can limit the number of .
Some QUPS functions use the currently selected parallel.gpu.CUDADevice, or select a parallel.gpu.CUDADevice by default. Use gpuDevice to manually select the gpu. Within a parallel pool, each worker can have a unique selection. By default, GPUs are spread evenly across all workers.
If CUDA support is enabled, ptx-files will be compiled to target the currently selected device. If the currently selected device changes, you may need to recompile binares using UltrasoundSystem.recompileCUDA or setup cache, particularly if the computer contains GPUs from different virtual architectures or have different compute capabilities.
Beamforming and simulation routines tend to require a trade-off between performance and memory usage. While QUPS attempts to balance this, you can still run into OOM errors if the GPU is almost full or if the problem is too large for either the GPU or CPU. There are several options to mitigate this such as:
- (GPU OOM) use
x = gather(x);to move some variables from the GPU (device) to the CPU (host) - this works onChannelDataobjects too. - for beamformers, set the
bsizeoptional keyword argument to a smaller value - (GPU OOM) use
gpuDevice().reset()to reset the currently selected GPU - NOTE: this will clear any variables that were not moved from the GPU to the CPU withgatherfirst - consider reducing the problem and using a for-loop e.g. by processing an image per frame or per depth, or simulating groups of 1000 scatterers at a time
- (GPU OOM) for
UltrasoundSystem.DASandUltrasoundSystem.greens, set thedeviceargument to0to avoid using a GPU device. - if a
parallel.Poolis active and a function accepts an optional parallel environment keyword argumentpenv, use a small number e.g.4to limit the number of workers, or0to avoid using the activeparallel.Pool - if a
parallel.Poolis active, shut it down withdelete(gcp('nocreate'))and disable automatic parallel pool creation.