Isoelectric point (pI) predictor for chemically modified peptides and proteins.
Published in:
Esben J. Bjerrum , Jan Holst Jensen, and Jakob L. Tolborg
J Chem Inf Model. 2017 Aug 28;57(8):1723-1727. doi: 10.1021/acs.jcim.7b00030.
Available at github.com/EBjerrum/pICalculax
A docker image has been compiled by Troels Schwarz-Linnet and is available at hub.docker.com/r/tlinnet/picalculax. Proteax Desktop is NOT installed in the image.
For handling condensed molfile formats, RDKit needs to be patched. Patch can be found in the rdkit_patch directory.
Patching RDKit can be difficucult. See the Dockerfile how this was done.
For handling conversion of protein line notation (PLN) to condensed molformat, Proteax desktop is needed. A modification database for Proteax Desktop import is found in the mods_db directory
Example usage of pICalculax can be found in the file Example_usage.py
The easiest solution, is to use the service mybinder.org, to launch an interactive Jupyter Notebook. Click here or the icon for access for online environment.
This can take up to 10 min, since the image is quite large.
Get prebuild image
docker pull tlinnet/picalculax:02_picalculaxRunning docker with image Link to run reference:
First make an alias
alias pi='docker run -ti --rm -p 8888:8888 -v "$PWD":/home/jovyan/work --name picalculax tlinnet/picalculax:02_picalculax'Run it
# With no arguments, starts Jupyter notebook
pi
# Or else start bash, to start programs
pi bashStart Docker image, with activated python 2.7 environment
$ pi pICalculax -h
Python 2.7.14 :: Anaconda custom (64-bit)
Calling pICalculax.py with arguments: -h
usage: pICalculax.py [-h] [--fasta FASTA [FASTA ...]] [--pln PLN [PLN ...]]
Predict isoeletric point pI of peptides and modified peptides
optional arguments:
-h, --help show this help message and exit
--fasta FASTA [FASTA ...]
Predict fasta sequence
--pln PLN [PLN ...] Predict PLN sequence (Requires Proteax Desktop)With FASTA
$ pi pICalculax --fasta ICECREAM FATCAT
Python 2.7.14 :: Anaconda custom (64-bit)
Calling pICalculax.py with arguments: --fasta ICECREAM FATCAT
4.14 ICECREAM
5.02 FATCATStart Docker image, with activated python 2.7 environment
pi py27from pICalculax import find_pKas, pI
from rdkit import Chem
fasta = 'ICECREAM'
mol = Chem.MolFromFASTA(fasta)
#find pKa values and charge class
pkalist, charge = find_pKas(mol)
#Calculate pI
pIpred = pI(pkalist, charge)
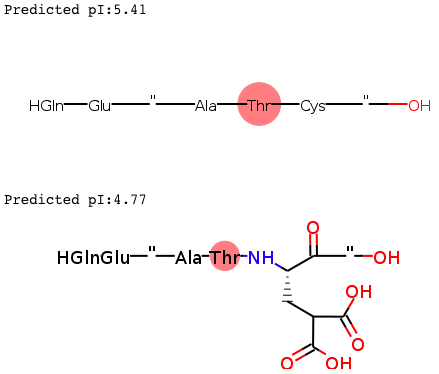
print(pIpred)Example usage of the pICalculax for pI prediction of unmodified and modified peptides
Start image with Jupyter notebook
piThen do as follow:
- In your browser, go to 0.0.0.0:8888
- Click New --> Python 2
- Paste in from below
The peptides can be loaded from a SDfile.
from pICalculax import find_pKas, pI
from rdkit import Chem
from rdkit.Chem import Draw
# https://github.com/rdkit/rdkit-tutorials/tree/master/notebooks
from rdkit.Chem.Draw import IPythonConsole
#Load a protein from SD file in condensed format
import os.path
paths = ['Datasets/example_mols.sdf', 'pICalculax_dir/Datasets/example_mols.sdf', '../pICalculax_dir/Datasets/example_mols.sdf']
for path in paths:
if os.path.exists(path):
sdsup = Chem.SDMolSupplier(path)
break
def predict_show(mol):
#Get list of identified pKa values and charge
pkalist, charge = find_pKas(mol)
#Predict the pI from the identified pKa values
piPred = pI(pkalist, charge)
#Report and Visualize
print("Predicted pI:%0.2F"%piPred)
# display is a Jupyter command
display(mol)
#Draw.ShowMol(mol, legend = "Predicted pI:%0.2F"%piPred)
#Draw.tkRoot.update()
#txt = raw_input('Press <ENTER> to continue')
# An unmodified peptide
mol = sdsup[0]
predict_show(mol)
# A peptide with a modification
mol = sdsup[1]
predict_show(mol)With Proteax Desktop protein line notation of modified peptides can be converted to a RDKit mol object and the pI predicted
from proteax_desktop import *
prtx = ProteaxDesktop()
pln = 'H-GHANY[Gla]A-OH'
mol = Chem.MolFromMolBlock(prtx.as_molfile(pln,'expansion-mode=condensed'))
#find pKa values and charge class
pkalist, charge = find_pKas(mol)
#Calculate pI
pIpred = pI(pkalist, charge)
print(pIpred)Protein line notation (Requires Proteax Desktop)
$ pi pICalculax --pln H-GHANYEA-OH H-GHANY[Gla]A-OH
5.41 H-GHANYEA-OH
4.77 H-GHANY[Gla]A-OH