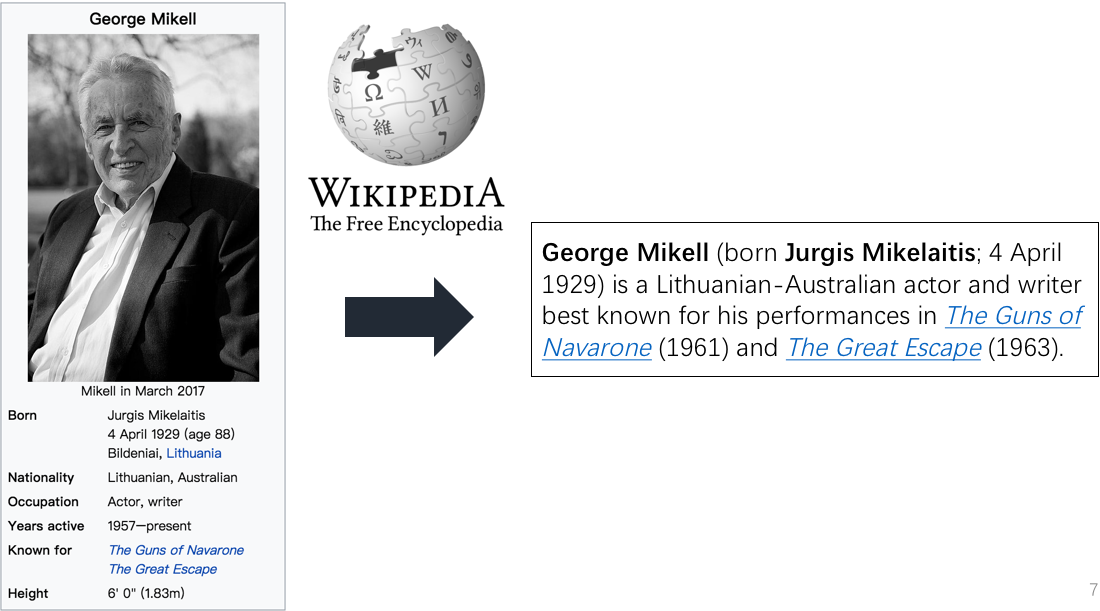
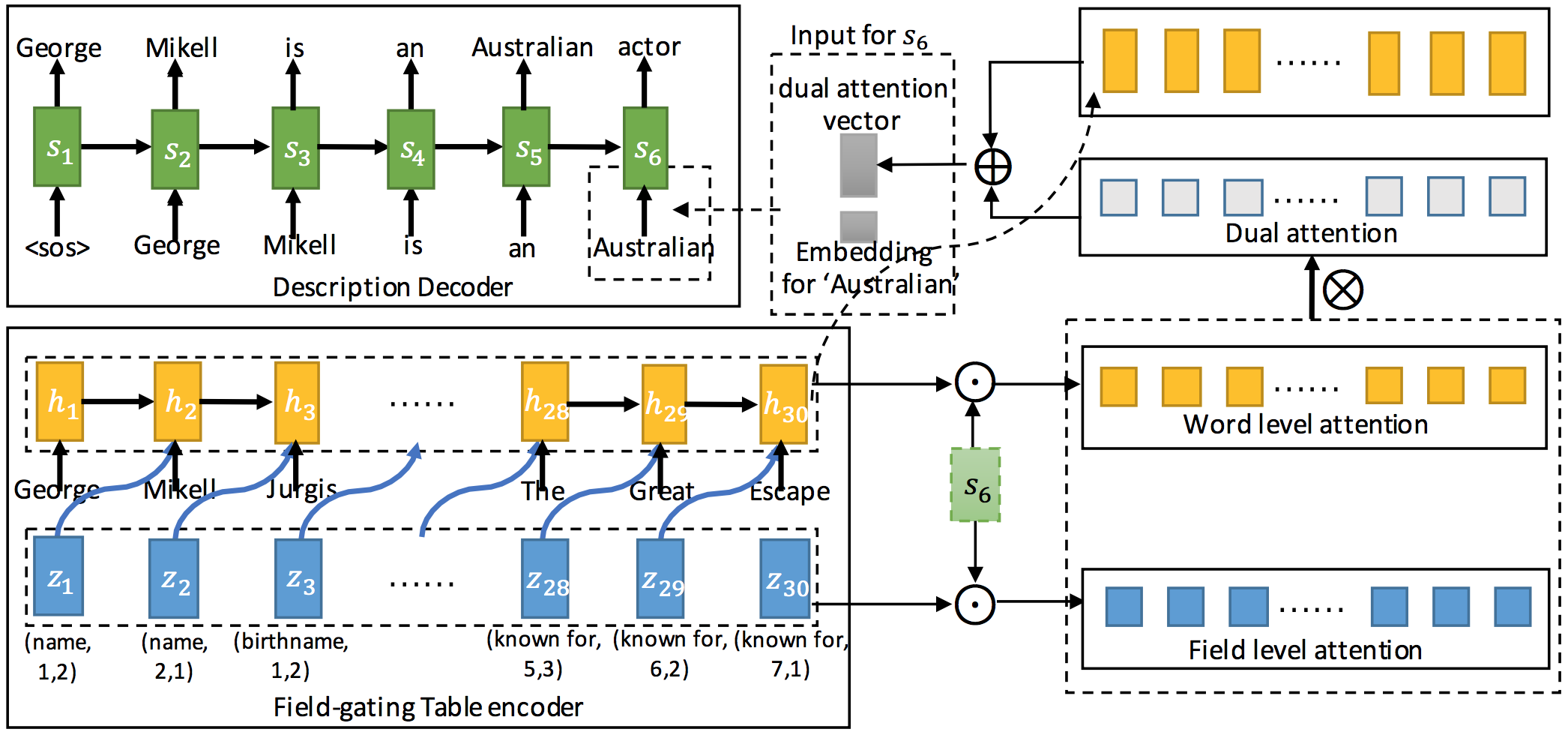
This project provides the implementation of table-to-text (infobox-to-biography) generation, taking the structure of a infobox for consideration.
Details of table-to-text generation can be found here. The implementation is based on Tensorflow 1.0.0 and Python 2.7.
wiki2bio is a natural language generation task which transforms Wikipedia infoboxes to corresponding biographies. We encode the structure of an infobox by taking field type and position information into consideration.In the encoding phase, we update the cell memory of the LSTM unit by a field gate and its corresponding field value in order to incorporate field information into table representation. In the decoding phase, dual attention mechanism which contains word level attention and field level attention is proposed to model the semantic relevance between the generated description and the table.
We strongly recommended using GPUs to train the model. It takes about 36~48 hours to finish training on a GTX1080 GPU.
Our code is based on Tensorflow 1.0.0. You can find the installation instructions here.
requirements.txt summarize the dependencies of our code. You can install these dependencies by:
pip install -r requirements.txt
The dataset for evaluation is WIKIBIO from Lebret et al. 2016. We preprocess the dataset in a easy-to-use way.
The original_data we proprocessed can be downloaded via Google Drive or Baidu Yunpan.
original_data
training set: train.box; train.summary
testing set: test.box; test.summary
valid set: valid.box; valid.summary
vocabularies: word_vocab.txt; field_vocab.txt
*.box in the original_data is the infoboxes from Wikipedia. One infobox per line.
*.summary in the original_data is the biographies corresponding to the infoboxes in *.box. One biography per line.
word_vocab.txt and field_vocab.txt are vocabularies for words (20000 words) and field types (1480 types), respectively.
The whole dataset is divided into training set (582,659 instances, 80%), valid set (72,831 instances, 10%) and testing set (72,831 instances, 10%).
Firstly, we extract words, field types and position information from the original infoboxes *.box.
After that, we idlize the extracted words and field type according to the word vocabulary word_vocab.txt and field vocabulary field_vocab.txt.
python preprocess.py
After preprocessing, the directory structure looks like follows:
-original_data
-processed_data
|-train
|-train.box.pos
|-train.box.rpos
|-train.box.val
|-train.box.lab
|-train.summary.id
|-train.box.val.id
|-train.box.lab.id
|-test
|-...
|-valid
|-...
-results
|-evaluation
|-res
*.box.pos, *.box.rpos, *.box.val, *.box.lab represents the word position p+, word position p-, field content and field types, respectively.
Experiment results will be stored in the results/res directory.
For training, turn the "mode" in main.py to train:
tf.app.flags.DEFINE_string("mode",'train','train or test')
Then run main.py:
python main.py
In the training stage, the model will report BLEU and ROUGE scores on the valid set and store the model parameters after certain training steps.
The detailed results will be stored in the results/res/CUR_MODEL_TIME_STAMP/log.txt.
For testing, turn the "mode" in main.py to train and the "load" to the selected model directory:
tf.app.flags.DEFINE_string("mode",'test','train or test')
tf.app.flags.DEFINE_string("load",'YOUR_BEST_MODEL_TIME_STAMP','load directory')
Then test your model by running:
python main.py
If you find the code and data resources helpful, please cite the following paper:
@article{liu2017table,
title={Table-to-text Generation by Structure-aware Seq2seq Learning},
author={Liu, Tianyu and Wang, Kexiang and Sha, Lei and Chang, Baobao and Sui, Zhifang},
journal={arXiv preprint arXiv:1711.09724},
year={2017}
}