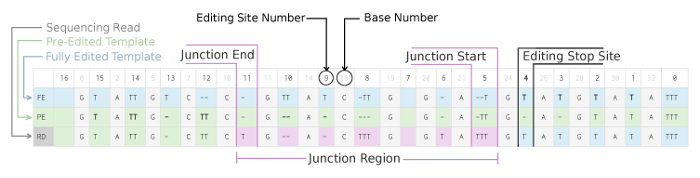
TREAT is a multiple sequence alignment and visualization tool specifically designed to analyze variation in sequences caused by Uridine insertion/deletion RNA editing. This phenomenon occurs in trypanosomes, a group of unicellular parasitic flagellate protozoa such the subspecies of Trypanosoma brucei which are the causative agents of Human African Trypanosomiasis (HAT or African Sleeping Sickness). The pre-mRNA sequences in trypanosomes are posttranscriptionally edited by the insertion/deletion of uridylate residues. TREAT aligns sequences using three bases and assembles editing sites to detect the extent of editing of the fourth base, called the edit base. The edit base is configurable in TREAT and by default uses 'T'. TREAT is written in Go and is released under the GPLv3 free software license.
Download the latest binary release for your system here. Extract the zip file:
$ unzip treat-0.0.x.zip $ cd treat-0.0.x $ ./treat --help
TREAT can perform a global alignment between two arbitrary sequences detecting the amount of insertion/deletions (indels) of a single base (called the "edit base"). The default edit base in TREAT is "T". For example:
$ ./treat align -1 ATCTGTATGT -2 ATTCGATTG -b T A-TCTGTA-TGT ATTC-G-ATTG-
TREAT can perform global alignments using template sequences. TREAT requires two user provided template sequences: fully edited and pre-edited. The fully edited template represents a mature edited mRNA transcript (completely precisely edited mRNA). The pre-edited template represents the sequence that will be edited in the mature RNA. TREAT accepts input sequences in FASTA format. For example:
# simple-templates.fa >Fully Edited CTTAATACACTTTTGATTAACAAACTTTAAA >Pre-Edited CTAATTACACTTTGATAACAAACTAAA # simple-sequences.fa >example-1 CTTAATTACACTTTGATTAACAAACTTTAAA
Save the above sequence files and run the alignment using TREAT. The example sequences can also be found in the ./examples directory:
$ ./treat align -t simple-templates.fa -f simple-sequences.fa ================================================================================ example-1 ================================================================================ JSS: 12 ESS: 11 JES: 18 Junc Len: 7 ================================================================================ FE: CTTAA-TACACTTTTGATTAACAAACTTTAAA PE: C-TAATTACAC-TTTGA-TAACAAAC--TAAA RD: CTTAATTACAC-TTTGATTAACAAACTTTAAA
TREAT computes the extent of canonical editing and reports various editing site characteristics as shown below:
TREAT can optionally store alignments into a database for more complex analysis, searching, and viewing in a web browser. TREAT has been tested on RNA-Seq data containing millions of sequences reads. Here's an example using sequences from ribosomal protein S12 (RPS12) from Trypanosoma brucei mitochondria.
TREAT accepts sequencing data in FASTA format. An example FASTA file (templates.fasta) containing the Fully Edited, Pre-Edited and one alternatively Edited template sequences is shown below:
>RPS12-FE Fully Edited CTAATACACTTTTGATAACAAACTAAAGTAAAtAtAttttGttttttttGCGtAtGtGAT TTTTGtAtGGttGttGtttACGttttGttttAtttGttttAtGttAttAtAtGAGtCCGC GAttGCCCAGttCCGGtAACCGACGtGtAttGtAtGCCGtAttttAttTAtAtAAttttG tttGGAtGttGCGttGttttttttGttGttttAttGGtttAGttAtGTCAttAtttAttA tAGAGGGTGGtGGttttGttGAtttACCCGGtGTAAAGtAttAtACACGTAttGtAAGtt AGATTTAGAtATAAGATATGTTTTT >RPS12-PE Pre-Edited CTAATACACTTTTGATAACAAACTAAAGTAAAAAGGCGAGGATTTTTTGAGTGGGACTGG AGAGAAAGAGCCGTTCGAGCCCAGCCGGAACCGACGGAGAGCTTCTTTTGAATAAAAGGG AGGCGGGGAGGAGAGTTTCAAAAAGATTTGGGTGGGGGGAACCCTTTGTTTTGGTTAAAG AAACATCGTTTAGAAGAGATTTTAGAATAAGATATGTTTTT >RPS12-A0 Alternative Editing (Cruz-Reyes 2013) alt_start=27 alt_stop=34 CTAATACACTTTTGATAACAAACTAAAGTAAAtAtAttttGttttttttGCGtAtGtGAT TTTTGtAtGGttGttGtttACGttttGttttAtttGttttAtGttAttAtAtGAGtCCGC GAttGCCCAGttCCGGtAACCGACGtGtAttGtAtGCCGtAttttAttTAtAtAAttttG tttGGAtGttGCGttGttttttttGttGttttAttGGtttAGttAtGTCAttAtttAttA tAGAGGGTGGtGGttttGttGAtttACCtCGttGGttTAtAtAGtAttAtACACGTAttG tAAGttAGATTTAGAtATAAGATATGTTTTT
FASTA file with our DNA fragment reads (sample-1.fasta):
>1-10 CTAATACACTTTTGATAACAAACTAAAGATATAATATTTTTGTTTTTTTTGCGTATGTGA TTTTTGTATGGTTGTTGTTTACGTTTTGTTTTATTTGTTTTATGTTATTATATGAGTCCG CGATTGCCCAGTTCCGGTAACCGACGTGTATTGTATGCCGTATTTTATTTATATAATTTT GTTTGGATGTTGCGTTGTTTTTTTTGTTGTTTTATTGGTTTAGTTATGTCATTATTTATT ATAGAGGGTGGTGGTTTTGTTGATTTACCCGGTGTAAAGTATTATACACGTATTGTAAGT TAGATTTAGATATAAGATATGTTTTT >2-9 CTAATACACTTTTGATAACAAACTAAAGTAAAAAGGCGAGGATTTTTTGAGTGGGATTCGGT ATTTGTTTTATGTTATTATATGAGTCCGCGATTGCCCAGCTCTGGTAACCGACGTGTATTGT ATGCCGTATTTTATTTATATAATTTTGTTTGGATGTTGCGTTGTTTTTTTTGTTGTTTTATT GGTTTAGTTATGTCATTATTTATTATAGAGGGTGGTGGTTTTGTTGATTTACCCGGTGTAAA GTATTATACACGTATTGTAAGTTAGATTTAGATATAAGATATGTTTTT >3-120 CTAATACACTTTTGATAACAAACTAAAGTAAAAAGGCGAGGATTTTTTGAGTGGGATTCGGTA TTTGTTTTATGTTATTATATGAGTCCGCGATTGCCCAGCTCTGGTAACCGACGTGTATTGTAT GCCGTATTTTATTTATATAATTTTGTTTGGATGTTGCGTTGTTTTTTTTGTTGTTTTATTGGT TTAGTTATGTCATTATTTATTATAGAGGGTGGTGGTTTTGTTGATTTACCCGGTGTAAAGTAT TATACACGTATTGTAAGTTAGATTTAGATATAACATATGTTTTT
Load the sample data using TREAT:
$ ./treat --db treat.db load --gene RPS12 \
--fasta sample-1.fa \
--template templates.fa \
--offset 10 \
--sample SampleName01 \
--knock-down GAP1 \
--tet \
--replicate 1
INFO[0000] Using template Edit Stop Site: 9
INFO[0000] Using Edit Site numbering offset: 10
INFO[0000] Processing fragments for sample name: SampleName01
INFO[0000] Done. Loaded 15 fragment sequences for sample SampleName01
A new database file has been created called "treat.db".
Normalize the read counts to 100000 (or an appropriate n) using the following command. Note: If you don't provide an n treat will normalize to the average read count across all samples within the gene:
$ ./treat --db testerino.db norm -n 100000 INFO[0000] Processing gene RPS12... INFO[0000] Normalizing to read count: 100000.0000 INFO[0000] Processing sample SampleName01 using normalized scaling factor: 9.3844
Search the data using the TREAT command line tool:
$ ./treat --db treat.db search -g RPS12 -l 10 --csv gene,sample,norm,read_count,alt_editing,has_mutation,edit_stop,junc_end,junc_len,junc_seq RPS12,sample-1,10.0000,10,0,0,137,143,6,ATATAATATTTTTG RPS12,sample-1,9.0000,9,0,0,95,123,28,TTCGGTATTTGTTTTATGTTATTATATGAGTCCGCGATTGCCCAGCTCTG
Search options are described below:
$ ./treat help search NAME: treat search - Search database USAGE: treat search [command options] [arguments...] OPTIONS: --gene, -g Gene Name --sample, -s [--sample option --sample option] One or more samples --edit-stop "-1" Edit stop --junc-end "-1" Junction end --junc-len "-1" Junction len --alt "0" Alt editing region --offset, -o "0" offset --limit, -l "0" limit --has-mutation Has mutation --all, -a Include all sequences --has-alt Has Alternative Editing --csv Output in csv format --fasta Output in fasta format --no-header, -x Exclude header from output
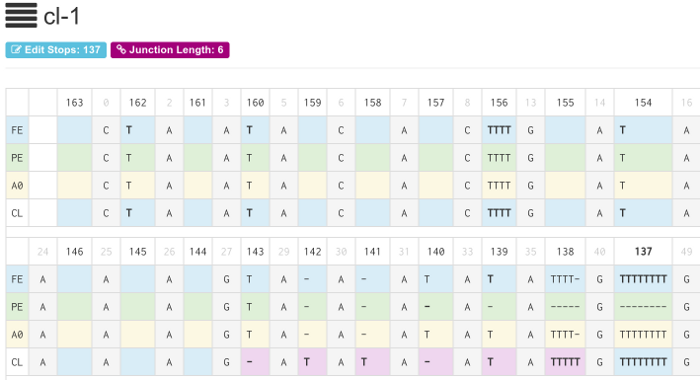
Start the TREAT server and view the sequences in a web browser:
$ ./treat --db treat.db server -p 8080 INFO[0000] Processing database: testerino.db INFO[0000] Computing cache for gene RPS12... INFO[0000] Max ESS: 21 Max JL: 12 Max JE: 21 INFO[0000] Using template dir: ./templates INFO[0000] Running on http://127.0.0.1:8080
To view the TREAT web interface, point your web browser at http://localhost:8080. By default, treat will listen on port 8080.
TREAT is written in Go requires v1.11 or greater. Clone the repository:
$ git clone https://github.com/ubccr/treat $ cd treat $ go build .
- Rachel M. Simpson, Andrew E. Bruno, Jonathan E. Bard, Michael J. Buck and Laurie K. Read. High-throughput sequencing of partially edited trypanosome mRNAs reveals barriers to editing progression and evidence for alternative editing. RNA, 2016. http://dx.doi.org/10.1261/rna.055160.115
TREAT is released under the GPLv3 license. See the LICENSE file. TREAT logo designed by Nicole B. Laski.