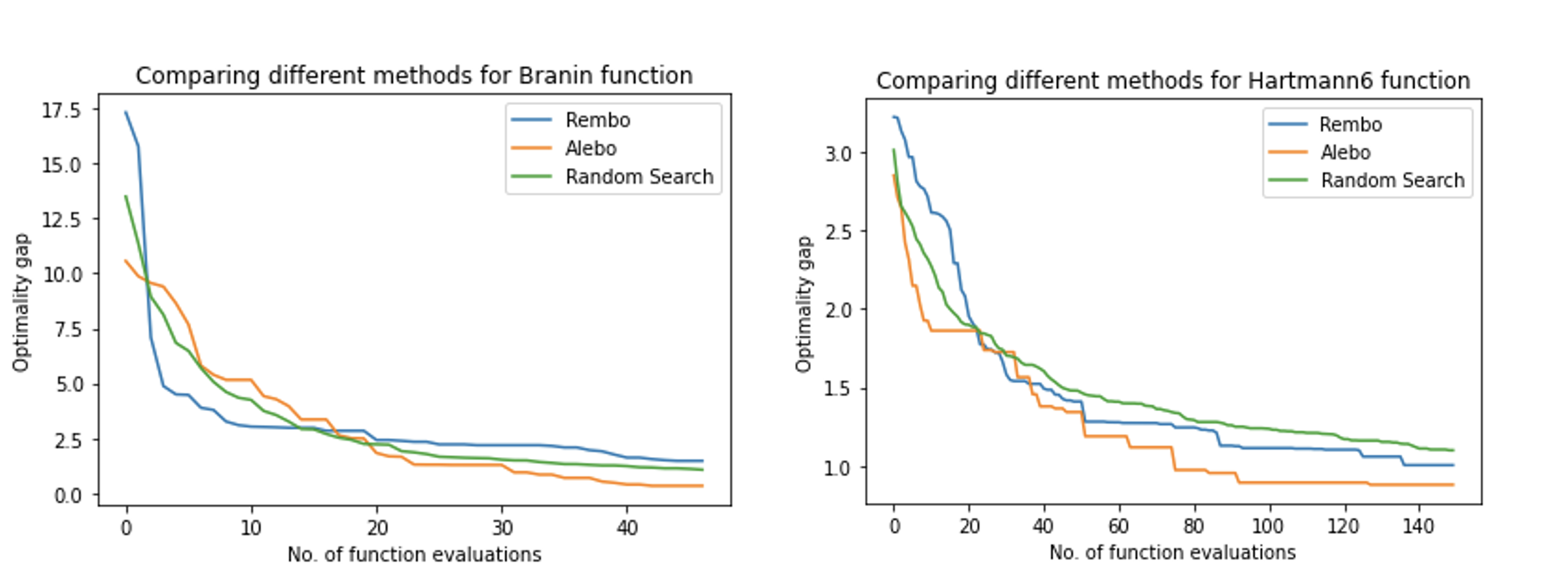
Implementation and review of techniques to scale bayesian optimization to high dimensions.
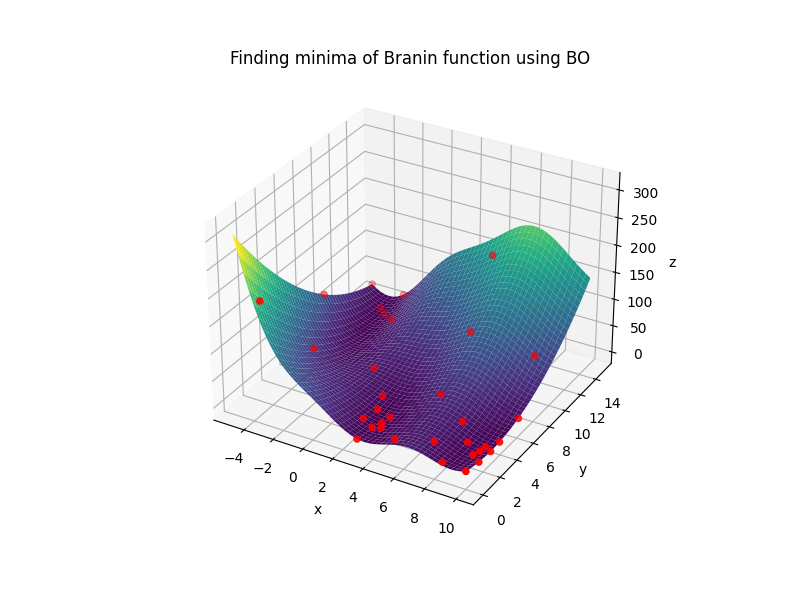
Example of running standard bayesian optimization on Branin function:
# Reference: https://www.sfu.ca/~ssurjano/branin.html
import pygpbo
import numpy as np
def F(p1,p2,noise=0):
return -(np.power((p2-5.1/(4*np.power(3.14,2))*np.power(p1,2)+5/3.14*p1-6),2)+10*(1-1/(8*3.14))*np.cos(p1)+10) + noise*np.random.randn()
bounds = {'p1': (-5, 10), 'p2': (0, 15)}
optimizer = pygpbo.BayesOpt(F, bounds)
optimizer.add_custom_points([{'p1': -4, 'p2': 1}, {'p1': -3, 'p2': 5},{'p1': 9, 'p2': 10},{'p1': 4, 'p2': 14}])
optimizer.maximize(n_iter=30)
print("Optimum point and value: ", optimizer.max_sample)
# Output seen: note that the optimum point can differ as branin has global minima at 3 different points
"""
Optimum point and value: ({'p1': 9.474934424992359, 'p2': 2.373669396349481}, array([-0.43209275]))
"""