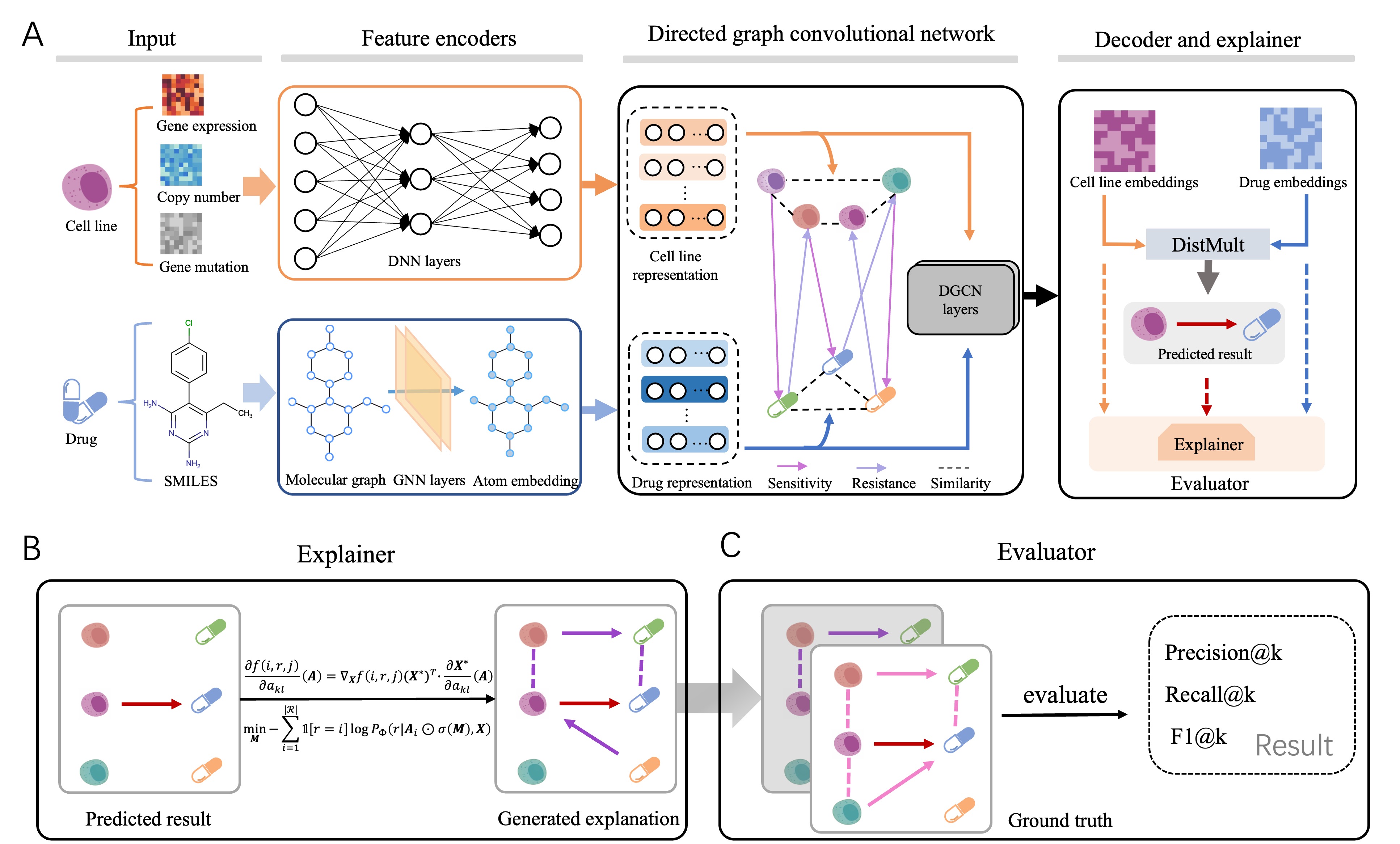
An official implement of Quantifiable Interpretability in Drug Response Prediction with Directed Graph Convolutional Network
To set up the required environment, you can choose from the following options:
-
Using conda: You can install the necessary Python dependencies from the
environment.ymlfile using the following command:conda env create -f environment.yml
-
/prediction - Contains all runnable code and data of prediction.
-
/explanation - Contains all runnable code and data of explanation.
python /prediction/code/DGCN.pypython /explanation/DRExplainer.pyHere is a list of the code files in this repository:
generate_feature.py- generates the feature of bio-entities and saves it to /prediction/data/node_representation.generate_negedge.py- generates the negative triples described in our paper and saves them to /prediction/data.DGCN.py- trains the prediction model and saves the model file to /prediction/data/weights.dgcn_eval.py- evaluates the model prediction performance in /prediction/data/weights.ground_truth.py- generates the ground truth benchmark dataset and save it to /explanation/data.DRExplainer.py- explaines the observable triples and save the results to /explanation/data.explain_eval.py- evaluates the model interpretability performance is /explanation/data.
If you need any help or are looking for cooperation feel free to contact us: haoyuan.shi@mail.ustc.edu.cn.