A package containing utility functions for the R-INLA package.
There's a fair bit of overlap with inlabru.
To install, first install INLA.
install.packages("INLA", repos="https://www.math.ntnu.no/inla/R/stable")then install INLAutils
# From github
library(devtools)
install_github('timcdlucas/INLAutils')
# Load packages
library(INLA)
library(INLAutils)Unfortunately, CRAN have now decided that as INLA is not on CRAN, INLAutils cannot be on CRAN either (a totally reasonable position).
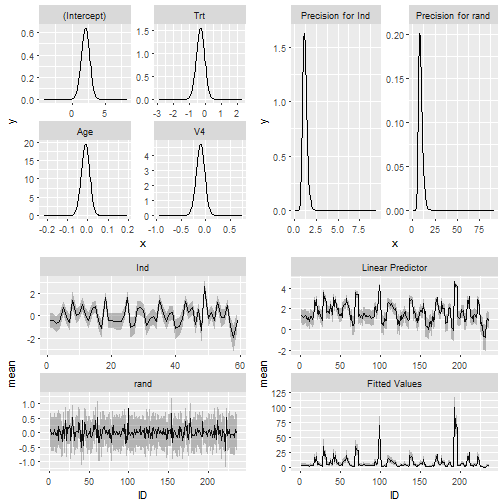
I find the the plot function in INLA annoying and I like ggplot2.
So INLAutils provides an autoplot method for INLA objects.
data(Epil)
##Define the model
formula = y ~ Trt + Age + V4 +
f(Ind, model="iid") + f(rand,model="iid")
result = inla(formula, family="poisson", data = Epil, control.predictor = list(compute = TRUE))
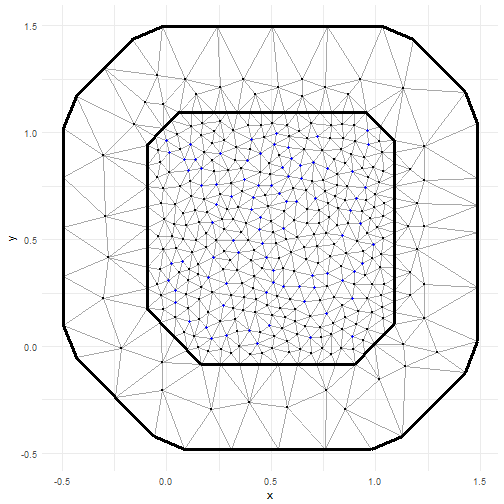
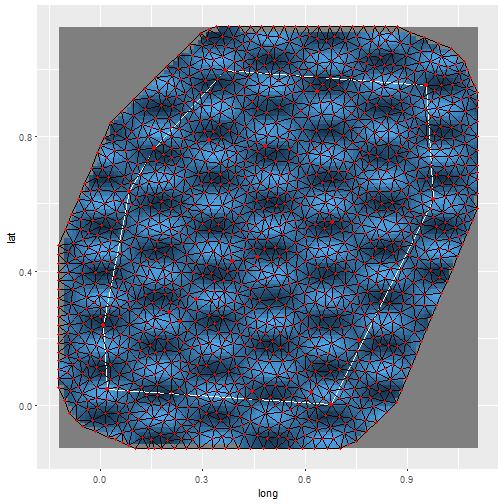
autoplot(result)There is an autoplot method for INLA SPDE meshes.
m = 100
points = matrix(runif(m * 2), m, 2)
mesh = inla.mesh.create.helper(
points = points,
cutoff = 0.05,
offset = c(0.1, 0.4),
max.edge = c(0.05, 0.5))
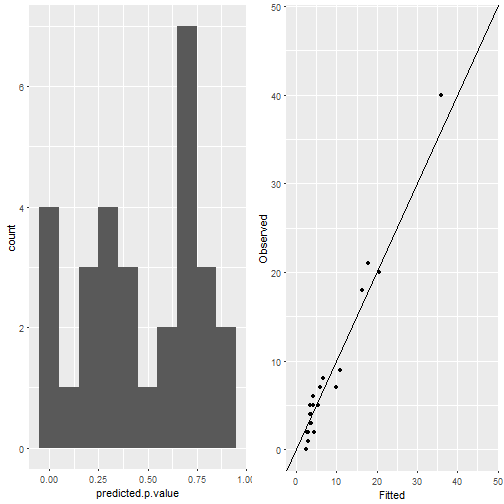
autoplot(mesh)There are functions for plotting more diagnostic plots.
data(Epil)
observed <- Epil[1:30, 'y']
Epil <- rbind(Epil, Epil[1:30, ])
Epil[1:30, 'y'] <- NA
## make centered covariates
formula = y ~ Trt + Age + V4 +
f(Ind, model="iid") + f(rand,model="iid")
result = inla(formula, family="poisson", data = Epil,
control.predictor = list(compute = TRUE, link = 1))
ggplot_inla_residuals(result, observed, binwidth = 0.1) ggplot_inla_residuals2(result, observed, se = FALSE)## `geom_smooth()` using method = 'loess'
Finally there is a function for combining shapefiles, rasters (or INLA projections) and meshes. For more fine grained control the geoms defined in inlabru might be useful.
# Create inla projector
n <- 20
loc <- matrix(runif(n*2), n, 2)
mesh <- inla.mesh.create(loc, refine=list(max.edge=0.05))
projector <- inla.mesh.projector(mesh)
field <- cos(mesh$loc[,1]*2*pi*3)*sin(mesh$loc[,2]*2*pi*7)
projection <- inla.mesh.project(projector, field)
# And a shape file
crds <- loc[chull(loc), ]
SPls <- SpatialPolygons(list(Polygons(list(Polygon(crds)), ID = 'a')))
# plot
ggplot_projection_shapefile(projection, projector, SPls, mesh)There are some helper functions for general analyses.
INLAstep runs stepwise variable selection with INLA.
data(Epil)
stack <- inla.stack(data = list(y = Epil$y),
A = list(1),
effects = list(data.frame(Intercept = 1, Epil[3:5])))
result <- INLAstep(fam1 = "poisson",
Epil,
in_stack = stack,
invariant = "0 + Intercept",
direction = 'backwards',
include = 3:5,
y = 'y',
y2 = 'y',
powerl = 1,
inter = 1,
thresh = 2)
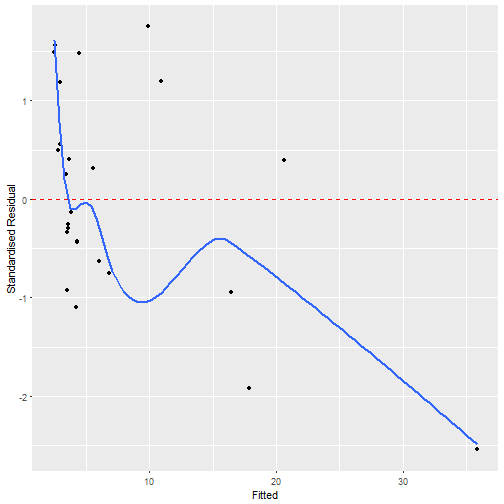
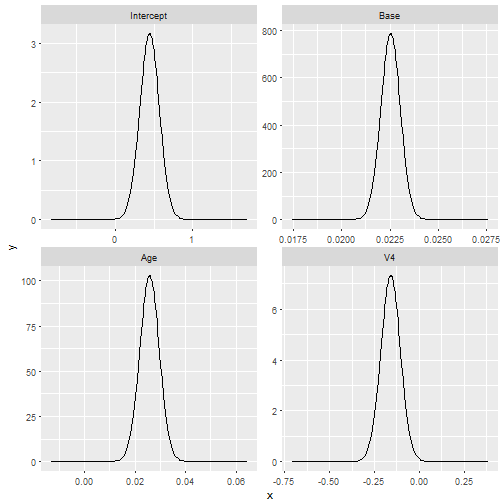
result$best_formula## y ~ 0 + Intercept + Base + Age + V4
## <environment: 0x0000000029d71cf0>
autoplot(result$best_model, which = 1)makeGAM helps create a function object for fitting GAMs with INLA.
data(Epil)
formula <- makeGAM('Age', invariant = '', linear = c('Age', 'Trt', 'V4'), returnstring = FALSE)
formula## y ~ +Age + Trt + V4 + f(inla.group(Age), model = "rw2")
## <environment: 0x000000001d543948>
result = inla(formula, family="poisson", data = Epil)inla.sdm- ggplot2 version of
plot.inla.tremesh - Make good
plotandggplotfunctions for plotting the Gaussian Random Field with value and uncertainty. stepINLA