This repo takes the output of Scott Grote's DREEM and adds additional data such as:
- per-sample information, such as the temperature or the cell line used.
- a library (per-construct information), such as the regions of interest in each construct (called sections), constructs families, etc.
- RNAstructure predictions for structure and free energy of each sequence.
- Poisson confidence intervals for mutation rates.
- RNAstructure (otherwise deactivate this option in the config file).
- Python packages described in
requirements.txt.
dreem-ppai is available on PyPi:
pip install dreem-ppai
You can also clone this repo and run:
cd path/to/where/you/want/dreem-ppai
git clone https://github.com/yvesmartindestaillades/dreem-ppai
cd dreem-ppai
python3 -m venv venv
source venv/bin/activate
pip install -r requirements.txt
Add RNAstructure path to test/config.yml:
gedit test/config.yml
Edit path:
# RNAstructure options
# ---------------------
rnastructure:
path: /Users/ymdt/src/RNAstructure/exe #where is RNAstructure installed
Then run:
python3 dreem-ppai/run.py
Download templates/config.yml.
If all of your csv files were processed on DREEM using the same fasta file, your csv files organization should look like this:
|- /[path_to_folder]
|- [your_sample_1].csv
|- [your_sample_2].csv
|- [your_sample_3].csv
|- [your_sample_4].csv
|- ...
If you used different fasta files, group your samples by mother fasta files and run dreem-ppai multiple times:
|- /[path_to_folder_1]
|- [your_sample_1].csv
|- [your_sample_2].csv
|- samples.csv
|- library_folder_1.csv
|- /[path_to_folder_2]
|- [your_sample_3].csv
|- [your_sample_4].csv
|- samples.csv
|- library_folder_2.csv
|- ...
Fill out this part of the template:
# Where to find your DREEM output files
# -------------------------------------
# This is the path to the directory where your DREEM output files are stored.
# The folder should be organized as follows:
# /path_to_dreem_output_files
# |- [your_sample_1].csv
# |- [your_sample_2].csv
# |- [your_sample_3].csv
# ...
path_to_dreem_output_files: /Users/ymdt/src/dreem-ppai/output_DREEM_mh
Fill out this part of the template:
# The samples that you want to process today
# ------------------------------------------
# These names must correspond to
# - the sample column in samples.csv
# - the name of your data folders
# Example:
# - [your_sample_1]
# - [your_sample_2]
# - [your_sample_3]
samples:
- 3UTR
- 5UTR
Fill out this part of the template:
# Where to store the results
# --------------------------
path_output: /Users/ymdt/src/dreem-ppai/output_DREEM_mh
dreem-ppai requires you to add the following mandatory informations for each samples, under the form of a csv file named samples.csv. Depending on exp_env, you also need to add buffer or cell_line.
all:
- sample # Sample name - CORRESPONDING TO THE NAME OF THE FASTQ FILE
- user # Who did the experiment
- date # Date of the experiment
- exp_env # Experimental environment, in_vivo or in_vitro
- temperature_k # Temperature en KelvIN
- inc_time_tot_secs # Total incubation time in seconds
- DMS_conc_mM # Concentration of DMS in mM
in_vitro:
- buffer # Exact buffer including Mg, eg 300mM Sodium Cacodylate , 3mM Mg
in_vivo:
- cell_line # Cell line
Download templates/samples.csv.
The template looks like this:
| sample | user | date | exp_env | temperature_k | inc_time_tot_secs | DMS_conc_mM | buffer | option1 | option2 |
|---|---|---|---|---|---|---|---|---|---|
Add columns to the csv file such as option1 and option2 in the template above.
|- /[path_to_folder]
|- samples.csv
|- [your_sample_1].csv
|- [your_sample_2].csv
|- [your_sample_3].csv
|- [your_sample_4].csv
|- ...
# Add info: add the uncommented lines to your DREEM outputs files
# --------------------------------------------------------------------
use:
samples: True # Add the content of samples.csv
...
Download templates/library.csv.
Library attributes are per-construct attributes, a construct being a name associated with a sequence, such as a line of your fasta file.
Examples:
- sections (regions of interest for this construct)
- sub-group (divide your constructs into sub-groups)
- barcode (enter the barcode associated with each construct)
Sections are defined by 3 columns:
- section_name
- section_start (1-indexed)
- section_stop (1-indexed, included)
When creating a section, all per-residue attributes (mutation rates, base coverage, etc) will be associated with the indexed defined by [section_start,section_stop]. If you want to have the full construct and also a section from this construct, you need to add a line for the construct without the section part.
/!\ Constructs that aren't in the library won't be saved in the output csv
library.csv
| construct | section_name | section_start | section_stop | is_region |
|---|---|---|---|---|
| my_construct_1 | not_a_region | |||
| my_construct_1 | MS2 | 19 | 42 | is_a_region |
| my_construct_1 | LAH | 67 | 81 | is_a_region |
| my_construct_2 | not_a_region | |||
| my_construct_3 | LAH | 73 | 90 | is_a_region |
| my_construct_3 | MS2 | 19 | 42 | is_a_region |
output.csv
| sample | construct | section_name | mut_rates | cov_bases | is_region |
|---|---|---|---|---|---|
| my_sample | my_construct_1 | full | [all mut rates] | [all cov_bases] | not_a_region |
| my_sample | my_construct_1 | MS2 | [mut rates for MS2] | [cov_bases for MS2] | is_a_region |
| my_sample | my_construct_1 | LAH | [mut rates for LAH] | [cov bases for LAH] | is_a_region |
| my_sample | my_construct_2 | full | [all mut rates] | [all cov_bases] | not_a_region |
| my_sample | my_construct_3 | LAH | [mut rates for LAH] | [cov bases for LAH] | is_a_region |
| my_sample | my_construct_3 | MS2 | [mut rates for MS2] | [cov_bases for MS2] | is_a_region |
|- /[path_to_folder]
|- samples.csv
|- library.csv
|- [your_sample_1].csv
|- [your_sample_2].csv
|- [your_sample_3].csv
|- [your_sample_4].csv
|- ...
# Add info: add the uncommented lines to your DREEM outputs files
# --------------------------------------------------------------------
use:
samples: True # Add the content of samples.csv
library: True # Add the content of library.csv
...
RNAstructure predictions are:
- structure
- free energy (deltaG)
The predictions are be done using:
- with/without temperature (if option set to True in the config file)
- with/without DMS signal as a constraint (set upper/lowerbounds for mutation probability normalization in the config file)
# RNAstructure options
# ---------------------
rnastructure:
path: /Users/ymdt/src/RNAstructure/exe #where is RNAstructure installed
temperature: False # Use samples.csv col 'temperature_k' as an input for RNAstructure
suffix_fold_cmd: '' # Additional input to add to the RNAstructure 'Fold' command
# for using DMS signal as an input in the argument
max_paired_mut_rate: 0.01 # below this value, 0% of the bases are unpaired
min_unpaired_mut_rate: 0.05 # above this value, 100% of the bases are unpaired
max_process: 64 # the maximum number of simultaneous Python subprocess when running RNAstructure
# Add info: add the uncommented lines to your DREEM outputs files
# --------------------------------------------------------------------
use:
samples: True # Add the content of samples.csv
library: True # Add the content of library.csv
rnastructure: True # Add RNAstructure
...
RNAstructure is a software from Prof. Mathews' lab. It predicts the structure of a RNA molecule and its thermodynamic energy based on Turner rules.
This is used to compute a confidence interval for each mutation rate of the population average.
Method:
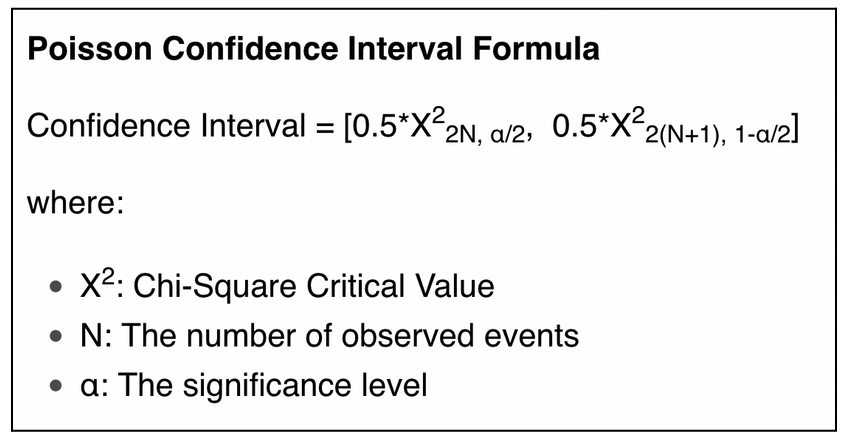
For each residue of a sequence, we model the probability of mutation by a binomial law. We approximate this binomial law by a Poisson distribution (Montgomery, 2001), and we use Poisson's confidence interval to compute a confidence interval for each residue of our population average.
The formula is the following:
A fully detailed document is available here.
# Add info: add the uncommented lines to your DREEM outputs files
# --------------------------------------------------------------------
use:
samples: True # Add the content of samples.csv
library: True # Add the content of library.csv
rnastructure: True # Add RNAstructure
poisson: True # Add Poisson confidence interval
python3
>>> from dreem-ppai import run
>>> run.run(config='config.yaml')
Export your pickle files to a csv or a json format by editing to_CSV or to_JSON in the config file.
Set verbose to True to get more informations in your terminal.
If you used different fasta files, group your samples by mother fasta files:
|- /[path_to_folder_1]
|- [your_sample_1].csv
|- [your_sample_2].csv
|- samples.csv
|- library_folder_1.csv
|- /[path_to_folder_2]
|- [your_sample_3].csv
|- [your_sample_4].csv
|- samples.csv
|- library_folder_2.csv
|- ...
Thanks for reading. Please contact me at yves@martin.yt for any additional information or to contribute.