This repo is a wrapper for Joe Yesselman's DREEM module, that implements the DREEM algorithm developed by the Rouskin lab.
The wrapper allows the user to run DREEM on different samples and to add standardized experimental details to DREEM output.
DREEM package must be in your environment. You need RNAstructure installed to run RNAstructure, otherwise deactivate this option in the config file.
dreem_herschlag is available on PyPi:
pip install dreem_herschlag
You can also clone this repo and run make:
cd [PATH_TO_WHERE_YOU_WANT_THE_REPO]
git clone https://github.com/yvesmartindestaillades/dreem_herschlag
cd dreem_herschlag
make init
If you want to use RNAstructure, open test/test_config.yml and assign dreem_args/RNAstructure_path to your path the RNAstructure/exe.
cd PATH/TO/REPO
dreem_herschlag --config test/test_config.yml
You should get this output:
$ dreem_herschlag --config test/test_config.yml
dreem_herschlag --config test/test_config.yml
Checking files
Checking test/resources/samples.csv
Checking samples.csv done
Checking case_1/library.csv
Ignored sequence, not in library_attributes
Checking case_1/library.csv done
Checking files done
Running DREEM
dreem -fq1 test/resources/case_1/r1.fastq -fq2 test/resources/case_1/r2.fastq -fa test/resources/case_1/ref.fasta --sample case_1 --sample_info temp/samples.csv --library_info temp/case_1/library.csv --overwrite
[19:27 bit_vector.py run] INFO ran at commandline as:
[19:27 bit_vector.py run] INFO /Users/ymdt/src/dreem_herschlag/bin/dreem -fq1 test/resources/case_1/r1.fastq -fq2 test/resources/case_1/r2.fastq -fa test/resources/case_1/ref.fasta --sample case_1 --sample_info temp/samples.csv --library_info temp/case_1/library.csv --overwrite
[19:27 bit_vector.py validate_files] INFO fasta file: test/resources/case_1/ref.fasta exists
[19:27 bit_vector.py validate_files] INFO fastq file: test/resources/case_1/r1.fastq exists
[19:27 bit_vector.py validate_files] INFO fastq2 file: test/resources/case_1/r2.fastq exists
[19:27 bit_vector.py validate_files] INFO two fastq files supplied, thus assuming paired reads
[19:27 bit_vector.py get_parameters] INFO -o/--overwrite supplied, will overwrite previous results with same name
[19:27 bit_vector.py build_directories] INFO building directory structure
[19:27 mapper.py __init__] INFO bowtie2 2.4.5 detected!
[19:27 mapper.py __init__] INFO fastqc v0.11.9 detected!
[19:27 mapper.py __init__] INFO trim_galore 0.6.6 detected!
[19:27 mapper.py __init__] INFO cutapt 1.18 detected!
[19:27 mapper.py __run_command] INFO running fastqc
[19:27 mapper.py __run_command] INFO fastqc ran without errors
[19:27 mapper.py __run_command] INFO running trim_galore
[19:27 mapper.py __run_command] INFO trim_galore ran without errors
[19:27 mapper.py __run_command] INFO running bowtie2-build
[19:27 mapper.py __run_command] INFO bowtie2-build ran without errors
[19:27 mapper.py __run_command] INFO running bowtie2 alignment
[19:27 mapper.py __run_command] INFO bowtie2 alignment ran without errors
[19:27 mapper.py __run_bowtie_alignment] INFO results for bowtie alignment:
2500 reads; of these:
2500 (100.00%) were paired; of these:
168 (6.72%) aligned concordantly 0 times
2331 (93.24%) aligned concordantly exactly 1 time
1 (0.04%) aligned concordantly >1 times
93.28% overall alignment rate
[19:27 mapper.py __run_picard_bam_convert] INFO Converting BAM file to SAM file format
[19:27 mapper.py __run_command] INFO running picard BAM conversion
[19:27 mapper.py __run_command] INFO picard BAM conversion ran without errors
[19:27 mapper.py __run_picard_sort] INFO sorting BAM file
[19:27 mapper.py __run_command] INFO running picard BAM sort
[19:27 mapper.py __run_command] INFO picard BAM sort ran without errors
[19:27 mapper.py run] INFO finished mapping!
[19:27 bit_vector.py run] INFO starting bitvector generation
[19:27 bit_vector.py __run_command] INFO running picard SAM convert
[19:27 bit_vector.py __run_command] INFO picard SAM convert ran without errors
[19:27 bit_vector.py run] INFO MUTATION SUMMARY:
| name | reads | aligned | no_mut | 1_mut | 2_mut | 3_mut | 3plus_mut | sn |
|---------------|---------|-----------|----------|---------|---------|---------|-------------|------|
| mttr-6-alt-h3 | 2332 | 99.96 | 46.42 | 36.81 | 13.21 | 3.05 | 0.04 | 7.76 |
None
DREEM done
This part will help you run DREEM with ease for multiple samples.
Your fasta/fastq files organization should look like this:
|- /[path_to_fastq_files]
|- samples.csv
|- library.csv
|- [some name].fasta
|- [your_sample_1]_R1_001.fastq.gz
|- [your_sample_1]_R2_001.fastq.gz
|- [your_sample_2]_R1_001.fastq.gz
|- [your_sample_2]_R2_001.fastq.gz
|- [your_sample_3]_R1_001.fastq.gz
|- [your_sample_3]_R2_001.fastq.gz
|- ...
We assume here that all smaples have the same fasta file and the same library. If you want to use different libraries, make several folders and run this module several times.`
- Download the
template_config.ymltemplate at the root of this repo, or generate it withdreem_herschlag --generate_templates - You may rename your file
my_config.ymlor whatever sounds good to you, so that you don't overwrite it. - Open the file and follow the fill-in instructions.
dreem_herschlag --config my_config.yml
This part will help you add additional content to your data:
- a library
library.csv, containing per-construct items. - a library
samples.csv, containing per-sample items. - various
RNAstructurepredictions, such as different structure and deltaG predictions and base-pairing probability. - binomial confidence intervals using the
Poissondistribution confidence interval.
Adding this content can be activated or deactivated in the config file.
Corresponding config file part:
# Add info mode: add the uncommented lines to your DREEM outputs files
# --------------------------------------------------------------------
add_info: False # turns this mode one
add_info_args:
library: True # Add the content of library.csv
samples: True # Add the content of samples.csv
rnastructure: True # Add RNAstructure
poisson: True # Add Poisson confidence interval
First, you have to create samples.csv and library.csv using the terminal.
TEMPLATES.CSV
Generate templates for samples.csv, library.csv and my_config.yml by running:
dreem_herschlag --generate_templates .
SAMPLES.CSV
Let's fill out samples.csv.
samples.csv contains information about each sample as a whole, such as the temperature or the date.
Each row of samples.csv correspond to a single sample.
The sample column of samples.csv must match the your_sample_# folders names shown above.
exp_env column content MUST BE set to in_vivo or in_vitro.
Columns description for samples.csv can be found by typing:
dreem_herschlag --sample_info
LIBRARY.CSV
Let's fill out library.csv.
library.csv contains information about each construct in a sample.
There must be one library.csv file per sample.
The name column of library.csv should match the constructs name of the fasta file.
Columns description for library.csv can be found by typing:
dreem_herschlag --library_info
RNAstructure is a software from Prof. Mathews' lab. It predicts the structure of a RNA molecule and its thermodynamic energy based on Turner rules.
RNAstructure predicts the following:
deltaG_min, i.e the energy of the most thermodynamically stable structure w.r.t Turner's rules.structure, i.e the most thermodynamically stable structure w.r.t Turner's rules.
You can add options in the config file to make addtional predictions:
dms, i.e RNAstructure uses the dms signal to predict the structure.temperature, i.e RNAstructure uses the temperature entered insamples.csvto predict the structure.roi, i.e RNAstructure makes predictions for the ROI defined inlibrary.csv.
The corresponding part in the config file is the following:
# RNAstructure options
# ---------------------
rnastructure:
temperature: False # Use samples.csv col 'temperature_k' as an input for RNAstructure
suffix_fold_cmd: '' # Additional input to add to the RNAstructure 'Fold' command
# for using DMS signal as an input in the argument
dms: True # Add predictions using DMS
max_paired_mut_rate: 0.01 # below this value, 0% of the bases are unpaired
min_unpaired_mut_rate: 0.05 # above this value, 100% of the bases are unpaired
temperature: True # Add predictions using temperature
roi: True # Add predictions using the ROI if there's a roi
Output attributes are the following:
deltaG, deltaG_DMS, deltaG_DMS_ROI, deltaG_DMS_ROI_T, deltaG_DMS_T, deltaG_ROI, deltaG_ROI_T, deltaG_T, deltaG_ens, deltaG_ens_ROI_ROI, deltaG_ens_ROI_ROI_T,deltaG_ens_T,mut_probability,structure,structure_DMS,structure_DMS_ROI,structure_DMS_ROI_T,structure_DMS_T,structure_ROI,structure_ROI_T,structure_T
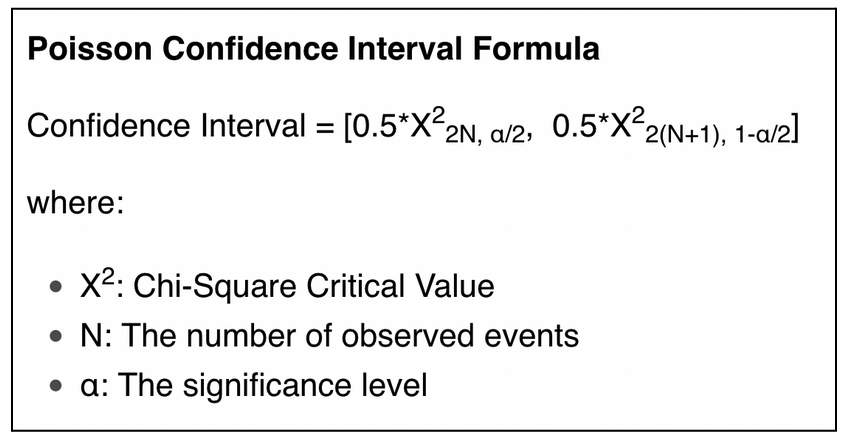
We want to get a confidence interval for each mutation rate of the population average.
This option can be activated in the config file, add_info_args/poisson: True.
Method:
For each residue of a sequence, we model the probability of mutation by a binomial law. We approximate this binomial law by a Poisson distribution (Montgomery, 2001), and we use Poisson's confidence interval to compute a confidence interval for each residue of our population average.
The formula is the following:
A fully detailed document is available here.
dreem_herschlag --config my_config.yml
Export your pickle files to a csv or a json format by editing to_CSV or to_JSON in the config file.
Set verbose to True to get more informations in your terminal.
Thanks for reading. Please contact me at yves@martin.yt for any additional information or to contribute.