Fast and simple parser for MEDLINE and Pubmed Open-Access affiliation string. We can parse multiple fields from the affiliation string including department, affiliation, location, country, email and zip code from affiliation text.
We also provide function to match affiliation string to GRID dataset.
Here is an example to parse affiliation
from affiliation_parser import parse_affil
parse_affil("Department of Health Science, Kochi Women's University, Kochi 780-8515, Japan. watanabe@cc.kochi-wu.ac.jp")output is a dictionary
{'full_text': "Department of Health Science, Kochi Women's University, Kochi , Japan. ",
'department': 'Department of Health Science',
'institution': "Kochi Women's University",
'location': 'Kochi , Japan',
'country': 'japan',
'zipcode': '780-8515',
'email': 'watanabe@cc.kochi-wu.ac.jp'}Here is an example to match affiliation to GRID dataset.
from affiliation_parser import match_affil
match_affil("Department of Health Science, Kochi Women's University, Kochi 780-8515, Japan. watanabe@cc.kochi-wu.ac.jp")Output is a dictionary consist of GRID ID
{'City': 'Kochi',
'Country': 'Japan',
'ID': 'grid.444150.0',
'Name': "Kochi Women's University",
'State': ''}use pip install -r requirements.txt in order to install required packages
Clone the repository and install using setup.py or simple copy affiliation_parser
folder to your workspace.
$ git clone https://github.com/titipata/affiliation_parser$ python setup.py installI put some snippet on how to produce quick summarization from MEDLINE data here.
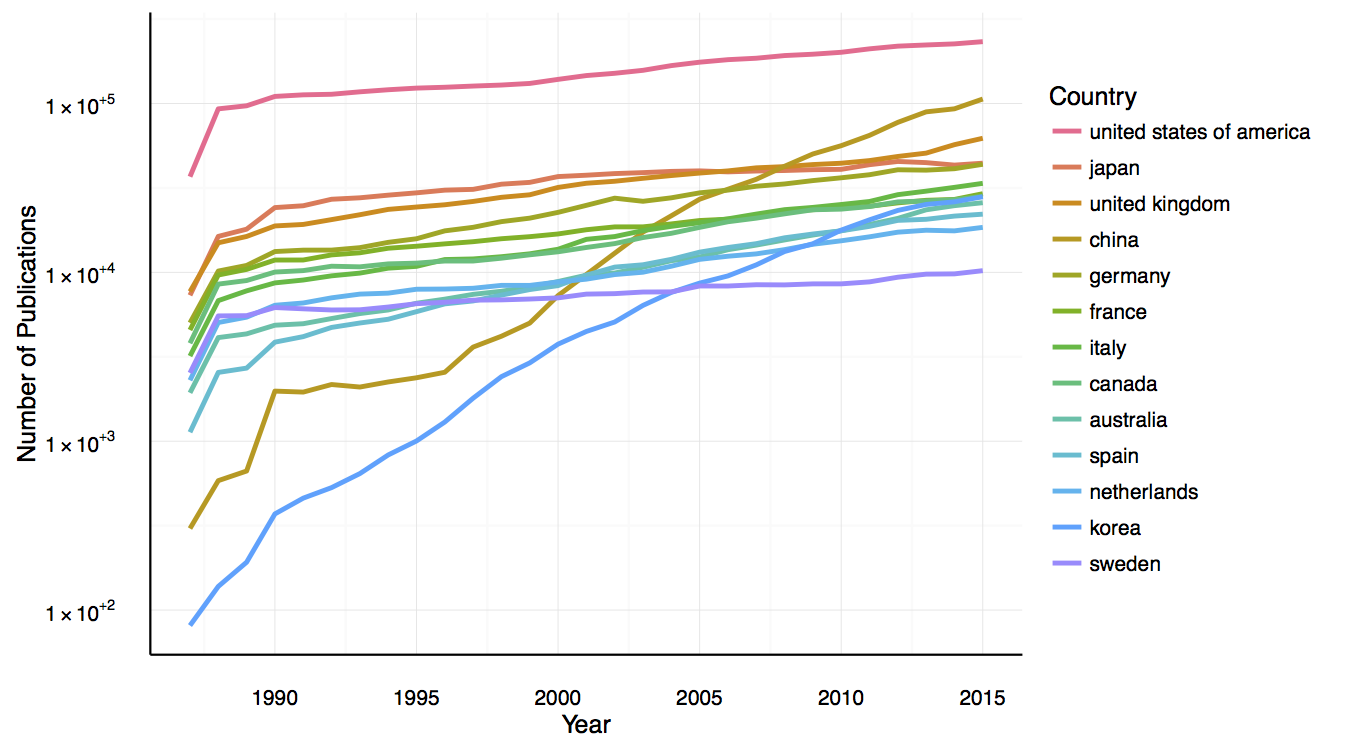
Total number of publications per country
Number of publications over time from selected countries