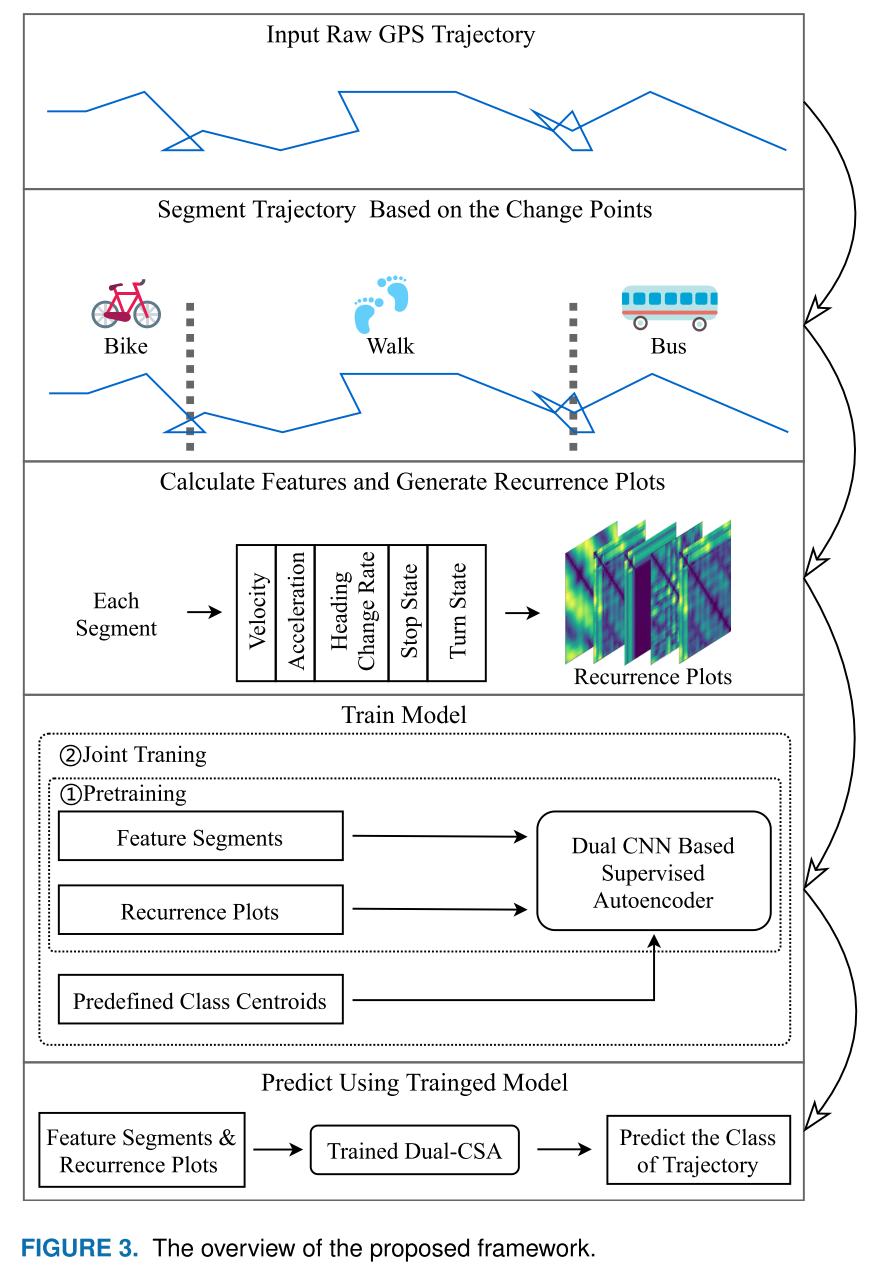
- Complete code support from preprocessing to results visualization. In particular, the preprocessing code is easy to migrate to other dataset.
- Most of the code is commented with detail explanation.
- The preprocessing support multi-process to speed up running time.
- The large data such as recurrence plots are generated in hard disk using Pytables, rather than RAM, which saves a lot of memory.
- The training support single GPU, multi-GPUs, multi-Nodes, and CPU.
- All the experiments in tha paper can be run using the shell scripts we wrote.
- Install PyTorch (pytorch.org)
pip install -r requirements.txt- Download Geolife dataset at: https://www.microsoft.com/en-us/research/project/geolife-buildingsocial-networks-using-human-location-history/#!downloads, then put all user folders under
Geolife Trajectories 1.3-Raw-All/Geolife Trajectories 1.3/Datainto our project folder./data/geolife_raw - Download SHL dataset at: http://www.shl-dataset.org/download/, then put all folders under
SHLDataset_User1Hips_v1/release/User1into our project folder./data/SHL_raw
- Extracted trajectories and labels from raw dataset & Generated handcrafted features and recurrence plots (RPs) are under
./data - Our main Dual-CSA network is under
network_torch/. network_variant/includes the variants of our Dual-CSA.ML_comparison/includes the classical machine learning methods we used to compare.network_comparison/includes other competitive deep learning based methods, note that they are implemented using keras.exp_scripts/includes all shell scripts we used to conduct experiments.keras_support_old/is the implementations of our model using keras, note we will not maintain keras version of our model any more.results/all training and predict results will be saved at this place.visualization_and_analysis/includes python code to draw some charts of experimental results in the paper.
- Before running the code, please set the environment variable of results path in the terminal, for example:
$ results_path=./results/exp1
$ export RES_PATH=${results_path}
- Please first run
trajectory_extraction_geolife.pyandtrajectory_extraction_SHL.pyto generate train & test trajectory and label.npyfile under./data/*_extracted. trajectory_segmentation_and_features_extraction.pyis used to segment the trajectory and extract movement features (MFs) and auxiliary features (AFs) for train or test set. For example:
python ./trajectory_segmentation_and_features_extraction.py --trjs_path ./data/geolife_extracted/trjs_train.npy --labels_path ./data/geolife_extracted/labels_train.npy --seg_size 200 --data_type train --save_dir ./data/geolife_features
where seg_size is max number of points in a segment.
MF_RP_mat_h5support.pyis used to generate RPs for feature segments generated above. For example:
python ./MF_RP_mat_h5support.py --dim 3 --tau 8 --multi_feature_segs_path ./data/geolife_features/multi_feature_segs_train.npy --save_path ./data/geolife_features/multi_channel_RP_mats_train.h5
where dim is embedding dimension & tau is the time delay in phase space reconstruction.
PEDCC.pyis used to generate the predefined evenly-distributed class centroids using the code of paper A Classification Supervised Auto-Encoder Based on Predefined Evenly-Distributed Class Centroids. For example:
python ./PEDCC.py --save_dir ./data/geolife_features --dim 304
where dim is embedding dimension in latent space.
network_training.pyis the training code of our model, our network can be trained on GPU, CPU, multi-GPU and multi-Nodes, its detail usage are:
usage: network_training.py [-h] --dataset DATASET [--network NETWORK]
[--results-path RESULTS_PATH]
[--RP-emb-dim RP_EMB_DIM] [--FS-emb-dim FS_EMB_DIM]
[--patience PATIENCE]
[--training-strategy TRAINING_STRATEGY]
[--no-save-model] [--visualize-emb VISUALIZE_EMB]
[--n-features N_FEATURES] [-j N]
[--pretrain-epochs N] [--joint-train-epochs N]
[-b N] [--wd W] [-p N] [--resume PATH] [-e]
[--pretrained] [--world-size WORLD_SIZE]
[--rank RANK] [--dist-url DIST_URL]
[--dist-backend DIST_BACKEND] [--seed SEED]
[--gpu GPU] [--multiprocessing-distributed]
DCSA_Training
optional arguments:
-h, --help show this help message and exit
--dataset DATASET geolife or SHL
--network NETWORK default is Dual_CSA, can be the variants: CSA-RP, CSA-
FS, and Dual-CA-Softmax
--results-path RESULTS_PATH
path to save the training and predict results
--RP-emb-dim RP_EMB_DIM
embedding dimension of RP autoencoder in latent space
--FS-emb-dim FS_EMB_DIM
embedding dimension of FS autoencoder in latent space
--patience PATIENCE patience of early stop in joint training
--training-strategy TRAINING_STRATEGY
can be: normal_training, normal_only_pretraining,
no_pre_joint_training, and only_joint_training
--no-save-model this flag will not save model
--visualize-emb VISUALIZE_EMB
this flag will turn on save latent visualization
images every visualize_emb epochs
--n-features N_FEATURES
number of MFs and AFs used, default 5
-j N, --workers N number of data loading workers (default: 4)
--pretrain-epochs N number of pretraining epochs to run
--joint-train-epochs N
number of joint training epochs to run
-b N, --batch-size N mini-batch size (default: 256), this is the total
batch size of all GPUs on the current node when using
Data Parallel or Distributed Data Parallel
--wd W, --weight-decay W
weight decay (default: 1e-4)
-p N, --print-freq N print frequency (default: 4)
--resume PATH path to latest checkpoint (default: none)
-e, --evaluate evaluate model on testation set
--pretrained use pre-trained model
--world-size WORLD_SIZE
number of nodes for distributed training
--rank RANK node rank for distributed training
--dist-url DIST_URL url used to set up distributed training
--dist-backend DIST_BACKEND
distributed backend
--seed SEED seed for initializing training.
--gpu GPU GPU id to use.
--multiprocessing-distributed
Use multi-processing distributed training to launch N
processes per node, which has N GPUs. This is the
fastest way to use PyTorch for either single node or
multi node data parallel training
Example usage for single nodes, one or multiple GPUs:
python network_training.py --dataset SHL --results-path ./results/exp1 --RP-emb-dim 152 --FS-emb-dim 152 --patience 20 --dist-url tcp://127.0.0.1:6666 --dist-backend nccl --multiprocessing-distributed --world-size 1 --rank 0 -b 230
Example usage for multiple nodes:
- Node 0:
python network_training.py --dataset SHL --results-path ./results/exp1 --RP-emb-dim 152 --FS-emb-dim 152 --patience 20 --dist-url tcp://127.0.0.1:6666 --dist-backend nccl --multiprocessing-distributed --world-size 2 --rank 0 -b 230
- Node 1:
python network_training.py --dataset SHL --results-path ./results/exp1 --RP-emb-dim 152 --FS-emb-dim 152 --patience 20 --dist-url tcp://127.0.0.1:6666 --dist-backend nccl --multiprocessing-distributed --world-size 2 --rank 0 -b 230
Example usage for CPU (Slow):
python network_training.py --dataset SHL --results-path ./results/exp1 --RP-emb-dim 152 --FS-emb-dim 152 --patience 20 -b 230
This is a shell script run under exp_scripts
cd ..
dataset='geolife'
results_path=./results/exp
export RES_PATH=${results_path}
python ./trajectory_segmentation_and_features_extraction.py --trjs_path ./data/SHL_extracted/trjs_train.npy --labels_path ./data/SHL_extracted/labels_train.npy --seg_size 200 --data_type train --save_dir ./data/SHL_features
python ./trajectory_segmentation_and_features_extraction.py --trjs_path ./data/SHL_extracted/trjs_test.npy --labels_path ./data/SHL_extracted/labels_test.npy --seg_size 200 --data_type test --save_dir ./data/SHL_features
python ./MF_RP_mat_h5support.py --dim 3 --tau 8 --multi_feature_segs_path ./data/SHL_features/multi_feature_segs_train.npy --save_path ./data/SHL_features/multi_channel_RP_mats_train.h5
python ./MF_RP_mat_h5support.py --dim 3 --tau 8 --multi_feature_segs_path ./data/SHL_features/multi_feature_segs_test.npy --save_path ./data/SHL_features/multi_channel_RP_mats_test.h5
python ./PEDCC.py --save_dir ./data/SHL_features --dim 304
python network_training.py --dataset ${dataset} --results-path ${results_path} --RP-emb-dim 152 --FS-emb-dim 152 --patience 20 --dist-url tcp://127.0.0.1:6666 --dist-backend nccl --multiprocessing-distributed --world-size 1 --rank 0 -b 230