This repo contains a proof of concept integration of FastZ with LASTZ version 1.04.06.
- nvidia cuda-toolkit 11.0
- cmake 3.8 or higher
- gcc and g++ 5.0 or higher
- python3
This code has only been tested on the plus strand using segment files that have been extracted from LASTZ.
make
./lastz ce11_chrIV.fa cb4_chrIV.fa --gapped --nogfextend --strand=plus --segments=chr4_first_two_mill_seed_subset --format=maf --output=chr4_first_two_mill_seed_subset.maf
python3 postprocess.py -i chr4_first_two_mill_seed_subset.maf
FastZ currently only supports maf files. An example maf file is included named chr4_first_two_mill_seed_subset.maf.
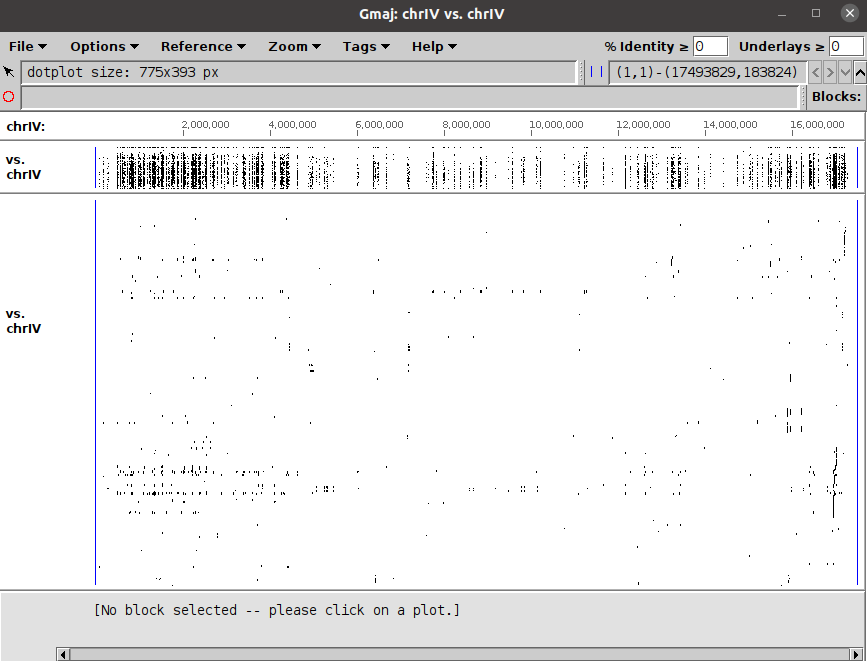
An opensource interactive software called Gmaj can be used to view the output files.
A preview of the sample maf file is as below.
Currently there are a defines in the src file FastZ.h which control various aspects of the code.
Important ones include.
MAXPROBSIZE - Largest alignment length observed across benchmarks, defaut = 30720
INSPSTREAMS - Number of streams across which the inspector kernel is launched, default = 32
INSTHREADS - Number of threads per inspector kernel launch, default = 128
INSPMEM - Float defining global memory for inspector, default 2.0 GB
The following tree shows the folder and file structures for the standalone implementation.
├── LICENSE
├── README.md
└── fastz
└── src
├── --default lastz src--
├── *.fa # test fasta files
├── ce11_chrIV.fa # example input fasta files
├── cb4_chrIV.fa # example input fasta files
├── chr4_first_two_mill_seed_subset.maf # example output file
├── FastZ.h
├── FastZ.cu
└── postprocess.py # post process script on the maf file
Sree Charan Gundabolu, T. N. Vijaykumar, and Mithuna Thottethodi. 2021. FastZ: accelerating gapped whole genome alignment on GPUs. In Proceedings of the International Conference for High Performance Computing, Networking, Storage and Analysis (SC '21). Association for Computing Machinery, New York, NY, USA, Article 8, 1–13. https://doi.org/10.1145/3458817.3476202