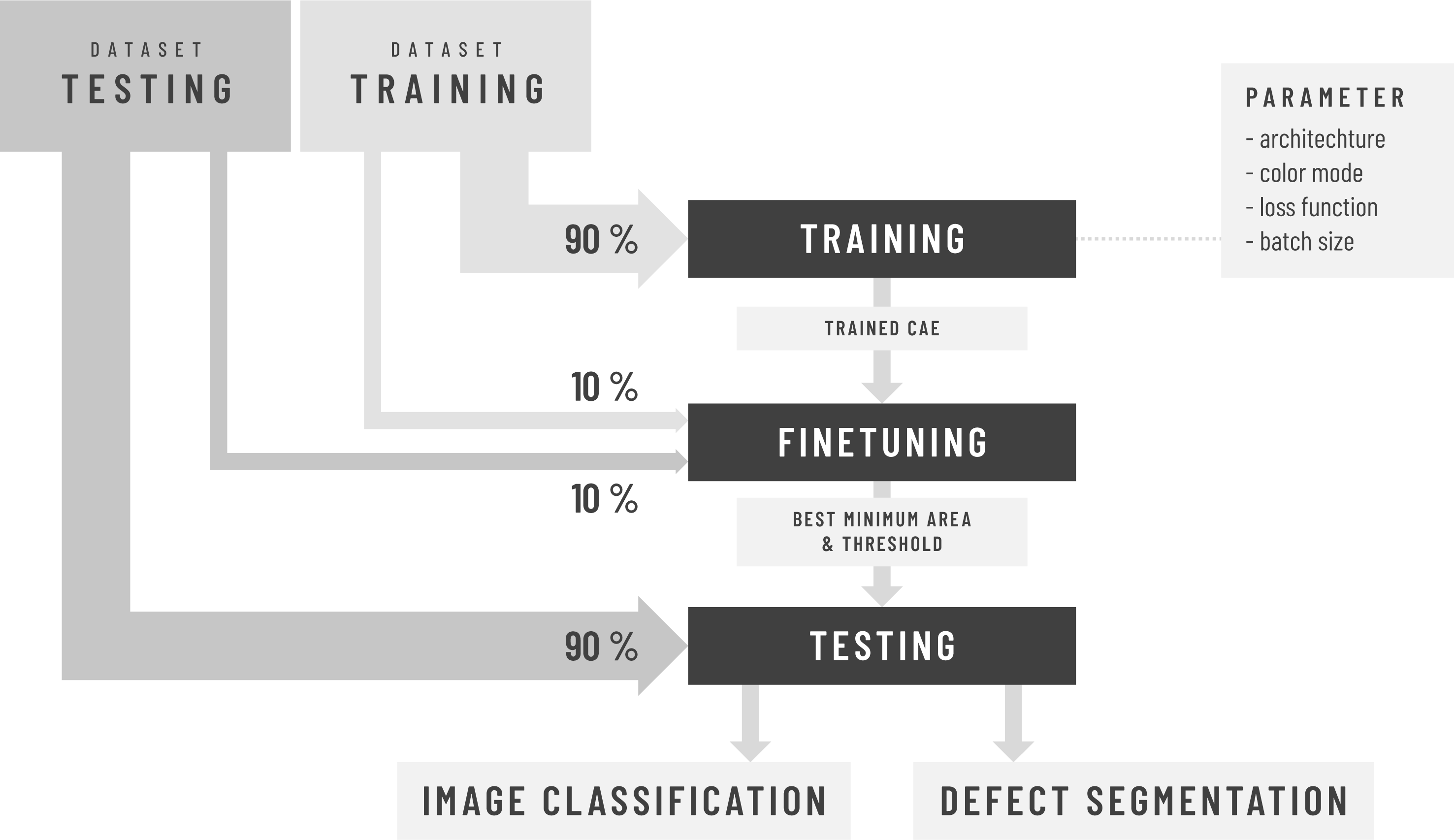
This project proposes an end-to-end framework for semi-supervised Anomaly Detection and Segmentation in images based on Deep Learning.
The proposed method employs a thresholded pixel-wise difference between reconstructed image and input image to localize anomaly. The threshold is determined by first using a subset of anomalous-free training images, i.e validation images, to determine possible values of minimum area and threshold pairs followed by using a subset of both anomalous-free and anomalous test images to select the best pair for classification and segmentation of the remaining test images.
It is inspired to a great extent by the papers MVTec AD — A Comprehensive Real-World Dataset for Unsupervised Anomaly Detection and Improving Unsupervised Defect Segmentation by Applying Structural Similarity to Autoencoders. The method is devided in 3 steps: training, finetuning and testing.
NOTE: Why Semi-Supervised and not Unsupervised?
The method proposed in the MVTec paper is unsupervised, as a subset containing only anomaly-free training images (validation set) are used during the validation step to determine the threshold for classification and segmentation of test images. However, the validation algorithm is based on a user input parameter, the minimum defect area, which definition remains unclear and unexplained in the aforementioned paper. Because the choice of this parameter can greatly influence the classification and segmentation results and in an effort to automate the process and remove the need for all user input, we developed a finetuning algorithm that computes different thresholds corresponding to a wide range of discrete minimum defect areas using the validation set. Subsequently, a small subset of anomaly and anomaly-free images of the test set (finetuning set) is used to select the best minimum defect area and threshold pait that will finally be used to classify and segment the remaining test images. Since our method relies on test images for finetuning, we describe it as being semi-supervised.
The proposed framework has been tested successfully on the MVTec dataset.
There is a total of 5 models based on the Convolutional Auto-Encoder (CAE) architecture implemented in this project:
- mvtecCAE is the model implemented in the MVTec Paper
- baselineCAE is inspired by: https://github.com/natasasdj/anomalyDetection
- inceptionCAE is inspired by: https://github.com/natasasdj/anomalyDetection
- resnetCAE is inspired by: https://arxiv.org/pdf/1606.08921.pdf
- skipCAE is inspired by: https://arxiv.org/pdf/1606.08921.pdf
NOTE:
mvtecCAE, baselineCAE and inceptionCAE are comparable in performance.
WARNING:
resnetCAE and skipCAE, are still being tested, as they are prone to overfitting, which translates in the case of convolutional auto-encoders by copying its inputs without filtering out the defective regions.
The main libraries used in this project with their corresponding versions are listed below:
tensorflow == 2.1.0ktrain == 0.21.3scikit-image == 0.16.2scikit-learn == 0.23.2
For more information, refer to requirement.txt.
Before installing dependencies, we highly recommend setting up a virtual anvironment (e.g., anaconda environment).
- Make sure pip is up-to-date with:
pip install -U pip - Install TensorFlow 2 if it is not already installed (e.g.,
pip install tensorflow==2.1). - Install ktrain:
pip install ktrain - Install scikit-image:
pip install scikit-image - Install scikit-learn:
pip install scikit-learn
The above should be all you need on Linux systems and cloud computing environments like Google Colab and AWS EC2. If you are using ktrain on a Windows computer, you can follow the more detailed instructions provided here that include some extra steps.
- Download the mvtec dataset here and save it to a directory of your choice (e.g in /Downloads)
- Extract the compressed image files.
- Create a folder in the project directory to store the image files.
- Move the extracted image files to that folder.
For the scripts to work propoerly, it is required for the folder containing the training and test images to have a specific structure. In the case of using the mvtec dataset, here is an example of how the directory stucture should look like:
├── bottle
│ ├── ground_truth
│ │ ├── broken_large
│ │ ├── broken_small
│ │ └── contamination
│ ├── test
│ │ ├── broken_large
│ │ ├── broken_small
│ │ ├── contamination
│ │ └── good
│ └── train
│ └── good
...
To train with your own dataset, you need to have a comparable directory structure. For example:
├── class1
│ ├── test
│ │ ├── good
│ │ ├── defect
│ └── train
│ └── good
├── class2
│ ├── test
│ │ ├── good
│ │ ├── defect
│ └── train
│ └── good
...
During training, the CAE trains exclusively on defect-free images and learns to reconstruct (predict) defect-free training samples.
usage: train.py [-h] -d [-a] [-c] [-l] [-b] [-i]
optional arguments:
-h, --help show this help message and exit
-d , --input-dir directory containing training images
-a , --architecture architecture of the model to use for training: 'mvtecCAE', 'baselineCAE', 'inceptionCAE' or 'resnetCAE'
-c , --color color mode for preprocessing images before training: 'rgb' or 'grayscale'
-l , --loss loss function to use for training: 'mssim', 'ssim' or 'l2'
-b , --batch batch size to use for training
-i, --inspect generate inspection plots after training
Example usage:
python3 train.py -d mvtec/capsule -a mvtecCAE -b 8 -l ssim -c grayscale
NOTE:
There is no need for the user to pass a number of epochs since the training process implements an Early Stopping strategy.
This script used a subset of defect-free training images and a subset of both defect and defect-free test images to determine good values for minimum defect area and threshold pair of parameters that will be used during testing for classification and segmentation.
usage: finetune.py [-h] -p [-m] [-t]
optional arguments: -h, --help show this help message and exit
-p , --path path to saved model
-m , --method method for generating resmaps: 'ssim' or 'l2'
-t , --dtype datatype for processing resmaps: 'float64' or 'uint8'
Example usage:
python3 finetune.py -p saved_models/mvtec/capsule/mvtecCAE/ssim/13-06-2020_15-35-10/mvtecCAE_b8_e39.hdf5 -m ssim -t float64
This script classifies test images using the minimum defect area and threshold previously approximated at the finetuning step.
usage: test.py [-h] -p [-s]
optional arguments: -h, --help show this help message and exit
-p , --path path to saved model
-s, --save save segmented images
Example usage:
python3 test.py -p saved_models/mvtec/capsule/mvtecCAE/ssim/13-06-2020_15-35-10/mvtecCAE_b8_e39.hdf5
├── mvtec <- folder containing all mvtec classes.
│ ├── bottle <- subfolder of a class (contains additional subfolders /train and /test).
| |── ...
├── autoencoder <- directory containing modules for training: autoencoder class and methods as well as custom losses and metrics.
├── processing <- directory containing modules for preprocessing images and before training and processing images after training.
├── results <- directory containing finetuning and test results.
├── readme.md <- readme file.
├── requirements.txt <- requirement text file containing used libraries.
├── saved_models <- directory containing saved models, training history, loss and learning plots and inspection images.
├── train.py <- training script to train the auto-encoder.
├── finetune.py <- approximates a good value for minimum area and threshold for classification.
└── test.py <- test script to classify images of the test set using finetuned parameters.
- Adnene Boumessouer - https://github.com/AdneneBoumessouer
This project is licensed under the MIT License - see the LICENSE.md file for details
- Paul Bergmann, Michael Fauser, David Sattlegger and Carsten Steger, the authors of the MVTec paper which this project has been inspired by.
- François Chollet, author of Keras.
- Aurélien Géron, autor of the great book Hands-On Machine Learning with Scikit-Learn, Keras, and TensorFlow, 2nd Edition.
- Arun S. Maiya, author of the ktrain deep learning library.
- Adrian Rosebrock, founder of the website pyimagesearch for computer vision.