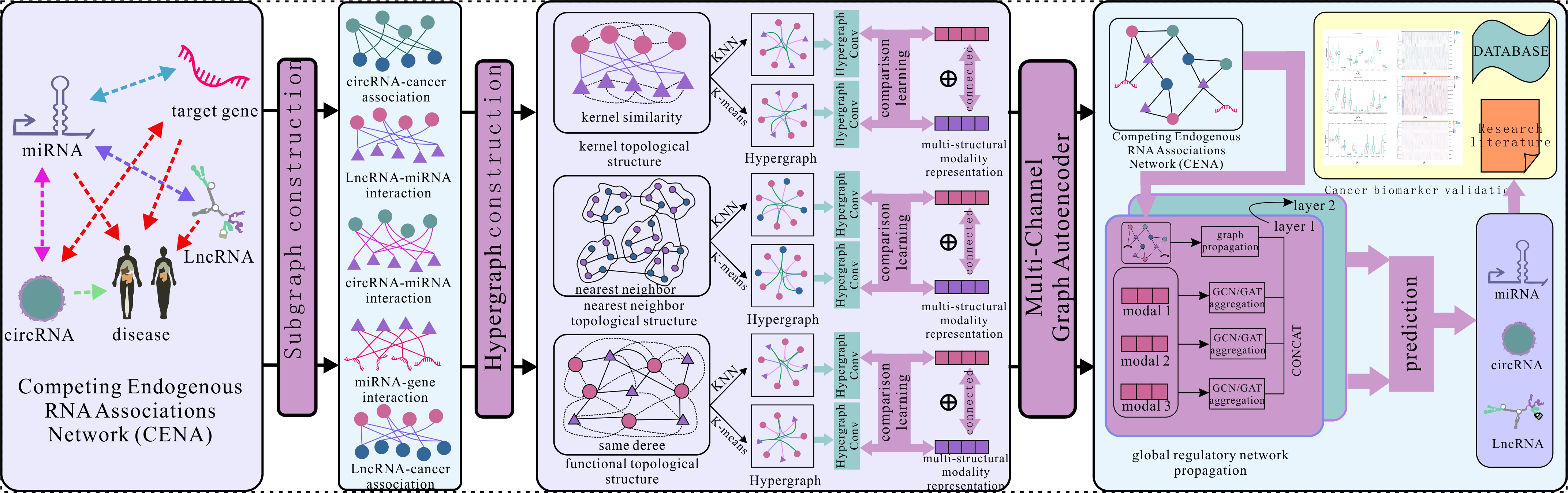
 MML-MGNN is an implementation of the
[A Multi-Channel Graph Neural Network based on Multi-Similarity Modality Hypergraph Contrastive Learning for Predicting Unknown Types of Cancer Biomarkers]
MML-MGNN is an implementation of the
[A Multi-Channel Graph Neural Network based on Multi-Similarity Modality Hypergraph Contrastive Learning for Predicting Unknown Types of Cancer Biomarkers]
The project includes data processing, experimental result presentation, and model training.
- Features
- Directory Structure
- Installation
- Usage
- Data
- Output
- Experimental Results
- mcgae
- Contributing
- License
- References
-Implementation of multi-similarity modal hypergraph contrastive learning (MMLMGN)
-Data processing and experimental workflow
-Showing experimental results in Jupyter Notebooks
-Using Python package tools for dependency management
-Implementation of prediction cases (mcgae)
MML-MGNN/
│
├── data/ # Example input files needed for running the code
│
├── experimental_results/ # All experimental results and associated Notebooks
│
├── mmlmgn/ # Contains all scripts and main.py for feature engineering
│
├── output/ # Output files generated by running main.py
│
├── mcgae/ # Example script to implement prediction
│
├── requirements.txt # Project dependencies
│
└── setup.py # Package configuration file
│
└── README.md # This document
To install the required dependencies in your environment, follow these steps:
-
Clone the repository:
git clone https://github.com/axin/MML-MGNN.git cd MML-MGNN -
Create a virtual environment:
python -m venv venv source venv/bin/activate -
Install the dependencies:
pip install -r requirements.txt
To run the main program, use the following command:
python mmlmgm/main.pyYou can pass command-line arguments when running main.py. Please refer to the code comments in param.py for details on the available arguments.
We are pleased to announce that the MMLMGN method for feature engineering has been packaged and uploaded to pypi.
You can install this package directly:
pip install MMLMGNFor detailed usage of MMLMGN, please refer to the readme description in pypi.
In the data/ folder, you can find example input files required for the code to run.
-
File
NodeGraph.csvis the graph input, used to construct the kernel similarity modal hyperedge of the node -
Files
NodeAWalker.csvandNodeBWalker.csvare the feature inputs of two nodes, with a size of N*M, n is the number of nodes, and M is the feature dimension, used to construct the nearest neighbor modal hyperedge -
Files
NodeASt.csvandNodeBSt.csvare the feature inputs of two nodes, with a size of N*M, n is the number of nodes, and M is the feature dimension, used to construct the structural topology modal hyperedge
Users can refer to data/ to customize input to use MMLMGN
In the output/ folder, you can find the three modality embedding files generated by the feature engineering MMLMGN for each node.
-
NodeAGraphEmb.csv,NodeBGraphEmb.csvare the kernel similarity modal embeddings of nodes A and B respectively. -
NodeAWalkerEmb.csv,NodeBWalkerEmb.csvare the nearest neighbor similarity modal embeddings of nodes A and B respectively. -
NodeAStEmb.csv,NodeBStEmb.csvare the structural topology similarity modal embeddings of nodes A and B respectively.
In the experimental_results/ folder, you can find detailed reports of all experimental results along with Jupyter Notebooks used for reproducing the figures. You can browse through these Notebooks to understand the model’s performance and effectiveness.
The experimental_results/ folder contains five subfolders, corresponding to the five comparative experiments in the results section of the paper.
- Contains the results of the benchmark data test (five-fold cross validation) for three cancer markers in the paper.
- CCA, LCA, MCA correspond to the circRNA, lncRNA, and miRNA biomarker prediction tasks for cancer, respectively.
- pre/test represents the model prediction score/data label.
- The numbers 0-4 represent the number of folds in the five-fold cross validation.
Visualization.ipynbImplemented the analysis of all the results in Cancer biomarker prediction and the reproduction of the charts in the paper.Visualization.ipynbis also applicable to the data indifferent aggregation layer/,different channel fusion/, andRobustness Testing/. You only need to change the path of the data.
- Results of different convolutional layer combinations on three benchmark tasks.
- Performance calculation and visualization are based on reference
Visualization.ipynb(the file directory needs to be changed). - The figures in the paper were drawn using Origin software after calculation in
Visualization.ipynb.
- Results of different channel fusion methods on three benchmark tasks.
- Performance calculation and visualization are based on reference
Visualization.ipynb(the file directory needs to be changed). - The figures in the paper were drawn using Origin software after calculation in
Visualization.ipynb.
- Results of robustness tests with different interference factors on three benchmark tasks.
- Performance calculation and visualization are based on reference
Visualization.ipynb(the file directory needs to be changed). - The figures in the paper were drawn using Origin software after calculation in
Visualization.ipynb.
-
Sota model comparison/ records the results of the SOTA model on three benchmark data sets (
9905/,9589/,CCA/) -
Three notebooks (
9905_SOTA.ipynb,9589_SOTA.ipynb,CCA_SOTA.ipynb) reproduce the figures comparing all models on the benchmark data.
-
mcgae provides an implementation of the multi-channel graph autoencoder used in the paper for downstream prediction tasks
-
mcgae aims to use these features to make predictions after feature engineering MMLMGN
-
mcgae shows how the npy file used to record the results is recorded
To run the mcgae program, use the following command:
python mcgae/MCGAE.pyContributions are welcome! If you would like to contribute to this project:
- Fork this repository
- Create your feature branch (
git checkout -b feature/YourFeature) - Commit your changes (
git commit -m 'Add some feature') - Push to the branch (
git push origin feature/YourFeature) - Open a pull request
This project is licensed under the MIT License. For more details, please see the LICENSE file.
Technical support provided by axin (xinfei106@gmail.com)