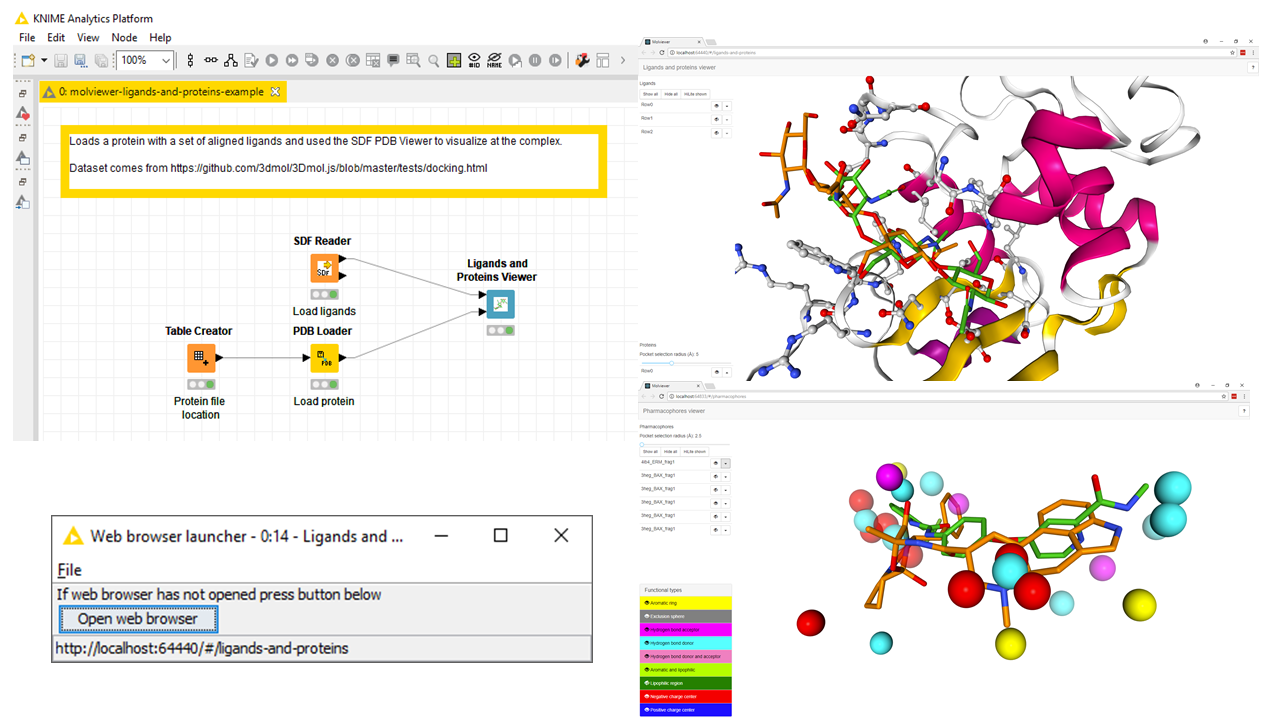
KNIME node which launches a web browser with a molecule viewer powered by NGL.
If you are using KNIME workflows with large molecules and you want to view them in 3D then the molviewer nodes are able to handle this.
- Provides cheminformatics trying to model proteins within KNIME a way to view them
- Adds nodes to KNIME to visualize proteins, small molecules and pharmacophores
- Adds support to KNIME to handle viewing big molecules
- Used to compare pharmacophore generation tools
- Used to check ligand repurposing
This project uses a web user interface based on https://github.com/3D-e-Chem/molviewer-tsx .
Installation
Requirements:
- KNIME, https://www.knime.org, version 4.0 or higher
Steps to get the MolViewer KNIME node inside KNIME:
- Goto Help > Install new software ... menu
- Press add button
- Fill text fields with
https://3d-e-chem.github.io/updates - Select --all sites-- in
work withpulldown - Select the node called
MolViewer nodes for KNIME - Install software
- Restart KNIME
Usage
-
Create a new KNIME workflow.
-
Find node in Node navigator panel under Community Nodes > 3D-e-Chem > Molviewer.
-
Drag node to workflow canvas.
-
Give molviewer nodes Proteins, Ligands and/or Pharmacophores as input.
-
Open the view of the molviewer node, this will launch a web browser with the molecules visualized.
-
In web browser you can
- Rotate/translate/zoom molecules with mouse
- Toggle which molecules are visible
- Transmit visible molecules as HiLite selection back to KNIME or do the reverse from KNIME to molviewer
- and more
See example workflow in examples/ directory.
Build
mvn verify
An Eclipse update site will be made in p2/target/repository repository.
The update site can be used to perform a local installation.
Development
Steps to get development environment setup based on https://github.com/knime/knime-sdk-setup#sdk-setup:
- Install Java 8
- Install Eclipse for RCP and RAP developers
- Configure Java 8 inside Eclipse Window > Preferences > Java > Installed JREs
- Import this repo as an Existing Maven project
- Activate target platform by going to Window > Preferences > Plug-in Development > Target Platform and check the
KNIME Analytics Platform (4.0) - nl.esciencecenter.e3dchem.knime.molviewer.targetplatform/KNIME-AP-4.0.targettarget definition. - A KNIME Analytics Platform instance can be started by right clicking on the
targetplatform/KNIME\ Analytics\ Platform.launchfile and selectingRun As → KNIME Analytics Platform. The KNIME instance will contain the target platform together with all extensions defined in the workspace.
During import the Tycho Eclipse providers must be installed.
Web interface
The web interface in the plugin/src/main/java/js-lib/molviewer directory. Is a distribution from the https://github.com/3D-e-Chem/molviewer-tsx repository.
Tests
Tests for the node are in tests/src directory.
Tests can be executed with mvn verify, they will be run in a separate KNIME environment.
Test results will be written to test/target/surefire-reports directory.
Workflow tests
See https://github.com/3D-e-Chem/knime-testflow#3-add-test-workflow
New release
- Update versions in pom files with
mvn org.eclipse.tycho:tycho-versions-plugin:set-version -DnewVersion=<version>-SNAPSHOTcommand. - Commit and push changes
- Create package with
mvn package, will create update site inp2/target/repository - Test node by installing it from local update site
- Append new release to an update site
- Make clone of an update site repo
- Append release to the update site with
mvn install -Dtarget.update.site=<path to update site> - Add files, commit and push changes of update site repo.
- Create a Github release
- Update Zenodo entry, fix authors
- Make nodes available to 3D-e-Chem KNIME feature by following steps at https://github.com/3D-e-Chem/knime-node-collection#new-release
- Update DOI in CITATION.cff
Technical architecture
The nodes are implemented as Dynamic JavaScript nodes also known as a org.knime.dynamic.node.generation.dynamicNodes extension.
The viewer has two way binding for the molecule visibility.
This project uses Eclipse Tycho to perform build steps.