A modification of the Prostate Segmenter library using our bone segmentation routines with CT images
- Video Demo of Bone Segmentation
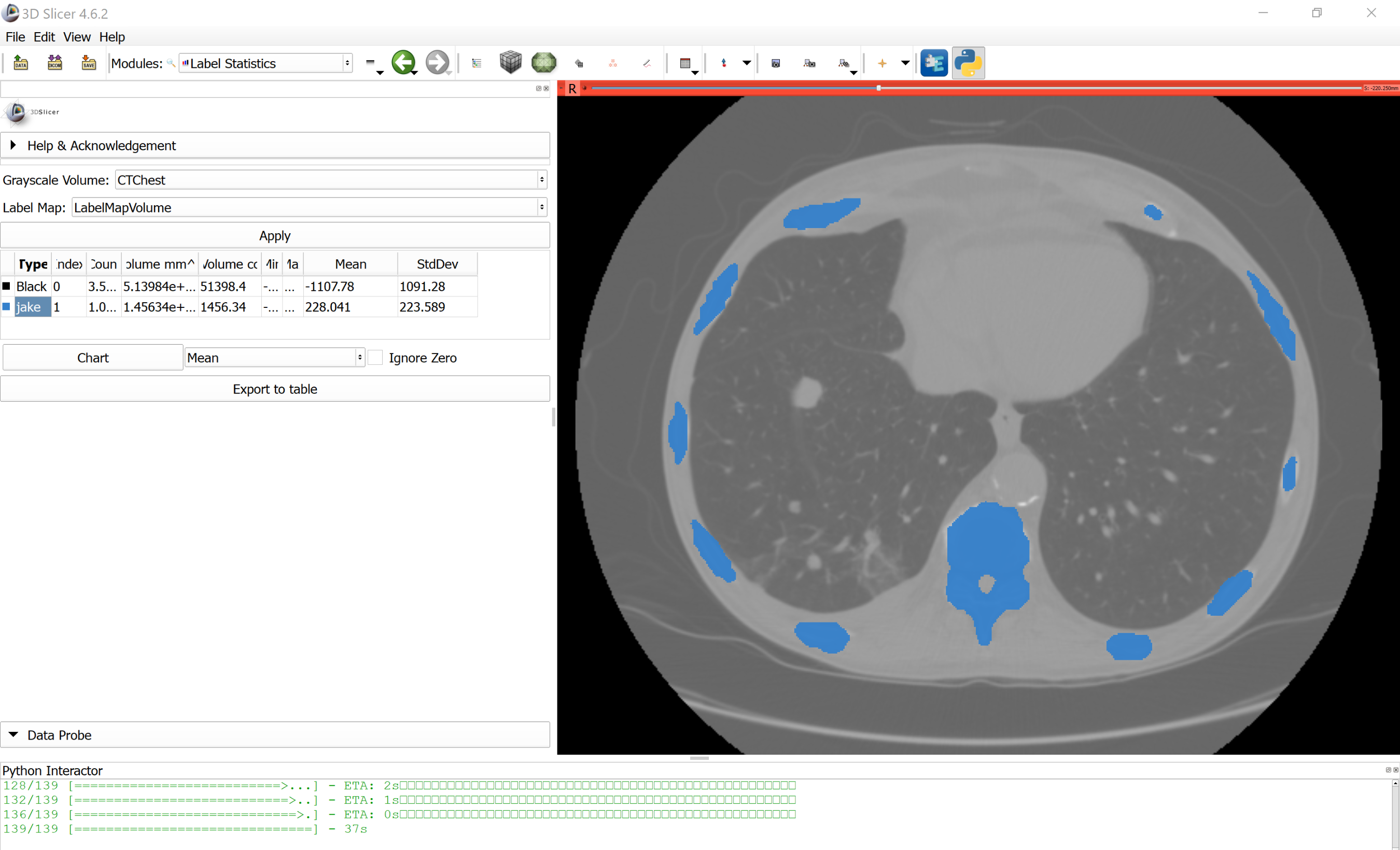
Using the DeepInfer plugin the image can be called interactively on a image loaded in Slicer. Here we show the results using the Chest CT image from the Sample Data
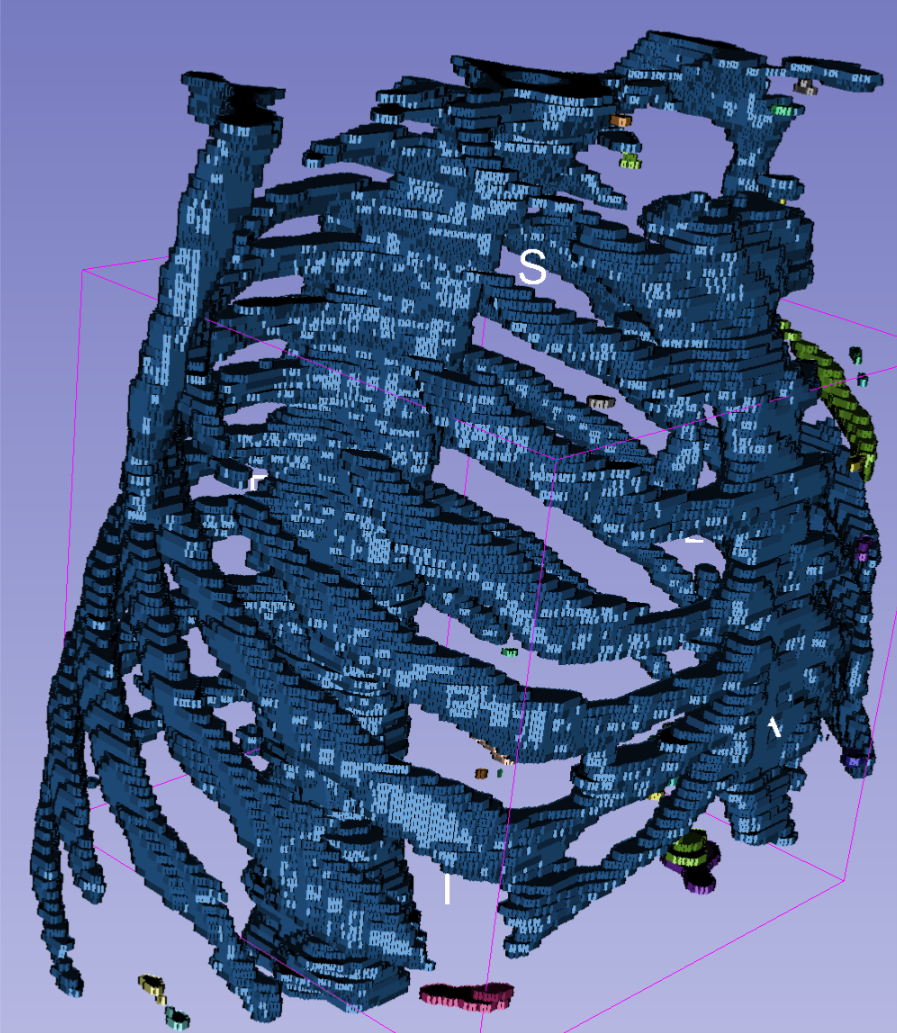
Here is then a 3D rendering of the segmented bones
docker pull 4Quant/bone-segmenter-cpu
The image and versions can also be found on the DockerHub site
You can use the following command to build the docker image again from the repository.
docker build -t 4quant/bone-segmenter-cpu -f Dockerfile.cpu .
docker run -t -v [Absolute PATH to the Project Folder]/Bone-Segmenter/data/test/:/home/deepinfer/data 4Quant/bone-segmenter-cpu --InputVolume /home/deepinfer/data/input.nrrd --OutputLabel /home/deepinfer/data/label_predicted_test.nrrd
Since paths in Windows are a bit different, you have to typically include the path in quotes which maps to the /home/deepinfer/data directory. It is also important to make sure Docker has access to the C drive.
docker run -t -v "C:\Users\Kevin Mader\Dropbox\4Quant\Projects\Bone-Segmenter":/home/deepinfer/data bone-segmenter-cpu --InputVolume /home/deepinfer/data/CTChest.nrrd --OutputLabel /home/deepinfer/data/label_predicted_test.nrrd
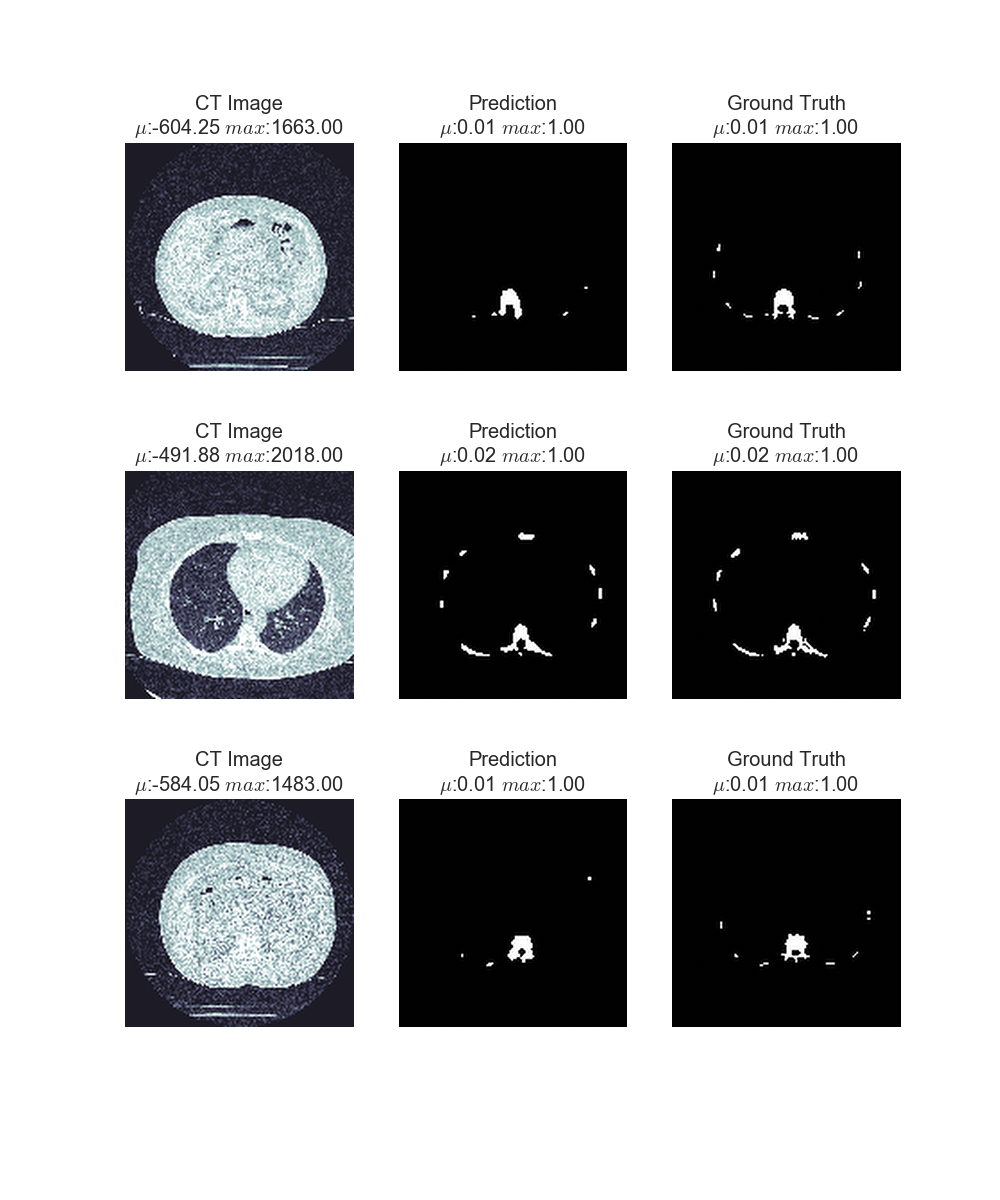
The predictions so two random images from the validation set including the predictions (middle column) by the network and the ground truth (right column). While the network does not always find smaller bones like ribs, it manages to accurately find vertebrae and hips.
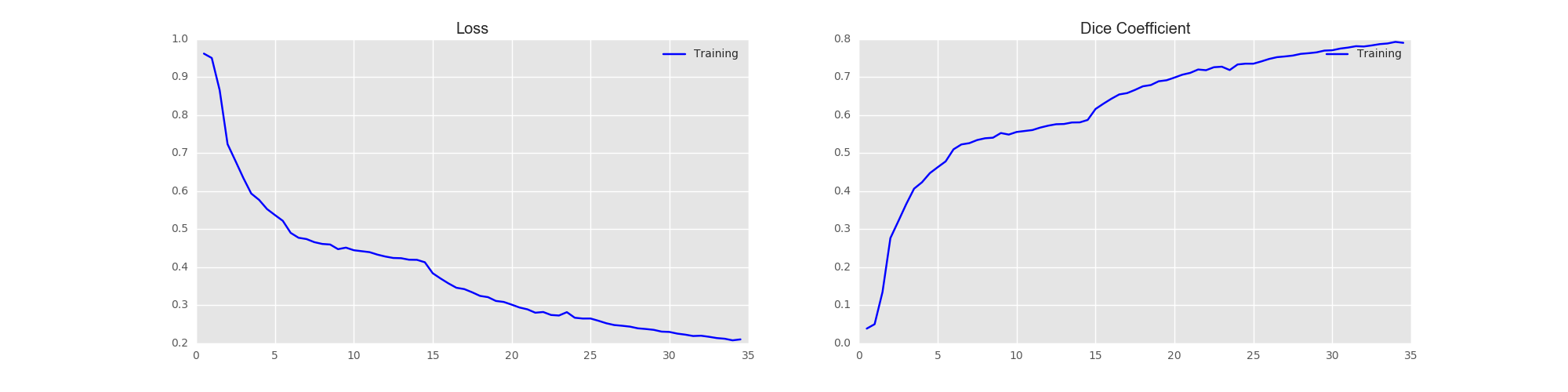
Here we show the training progress and corresponding DICE score.