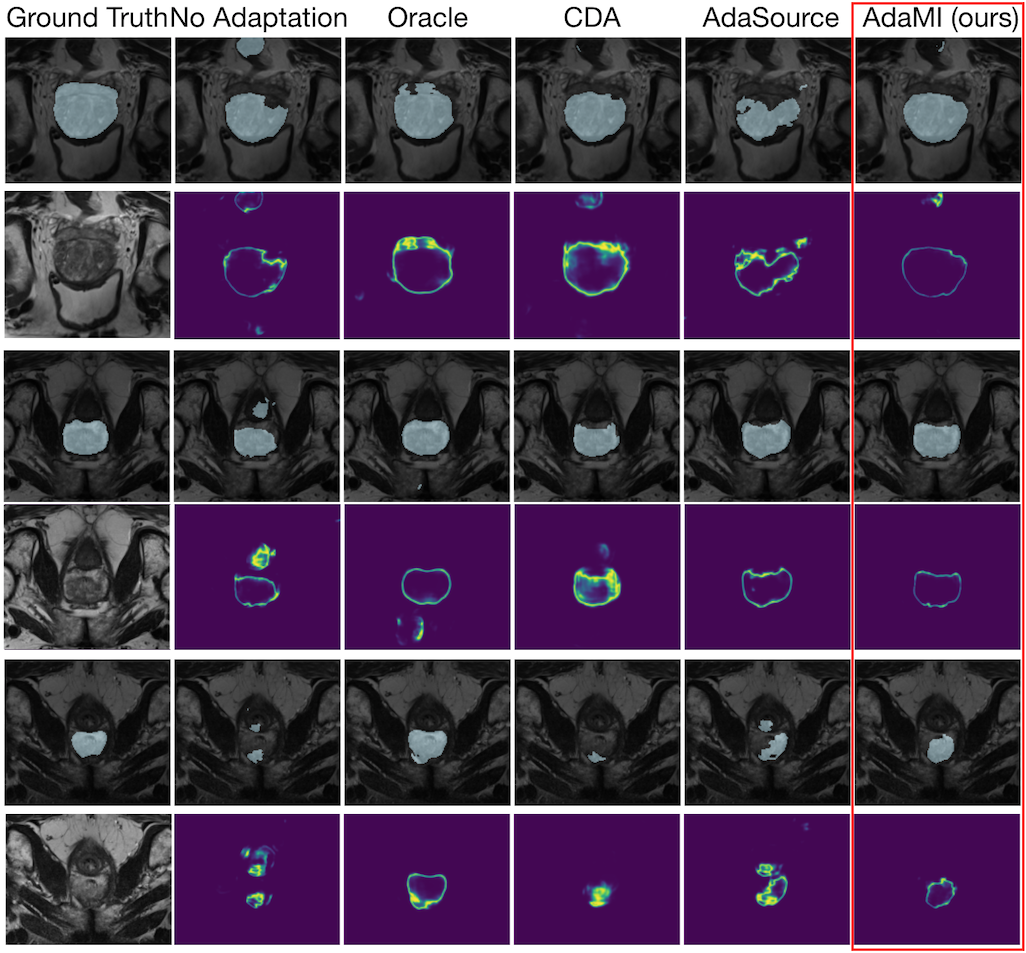
We introduce Source-Free Domain Adaptation for Image Segmentation.
Mathilde Bateson, Hoel Kervadec, Jose Dolz, Hervé Lombaert, Ismail Ben Ayed @ETS Montréal
Code of our submission at MICCAI 2020 and its ongoing journal extension. Video of the MICCAI talk is available: https://www.youtube.com/watch?v=ALYaa5xrxbQ&ab_channel=MB
Please cite our paper if you find it useful for your research.
@inproceedings{BatesonSFDA,
Author = {Bateson, Mathilde and Kervadec, Hoel and Dolz, Jose and Lombaert, Herv{\'e} and Ben Ayed, Ismail},
Booktitle = {Medical Image Computing and Computer Assisted Intervention -- MICCAI 2020},
Pages = {490--499},
Publisher = {Springer International Publishing},
Title = {Source-Relaxed Domain Adaptation for Image Segmentation},
Year = {2020}
Address = {Cham}}
Non-exhaustive list:
- python3.6+
- Pytorch 1.0
- nibabel
- Scipy
- NumPy
- Matplotlib
- Scikit-image
- zsh
- tqdm
- pandas
- scikit-image
For instance
data
prostate_source/
train/
IMG/
Case10_0.png
...
GT/
Case10_0.png
...
...
val/
IMG/
Case11_0.png
...
GT/
Case11_0.png
...
...
prostate_target/
train/
IMG/
Case10_0.png
...
GT/
Case10_0.png
...
...
val/
IMG/
Case11_0.png
...
GT/
Case11_0.png
...
...
The network takes png or nii files as an input. The gt folder contains gray-scale images of the ground-truth, where the gray-scale level is the number of the class (0,1,...K).
The class-ratio prior is estimated from anatomical knowledge for each application. In our implementation, is it estimated for each slice in the target domain training and validation sets. It estimated once, before the start of the adaptation phase, and saved in a csv file.
Scheme
sizes/
prostate.csv
whs.csv
ivd.csv
The size csv file should be organized as follows:
| val_ids | dumbpredwtags |
|---|---|
| Case00_0.nii | [Estimated_Size_class0, Estimated_Size_class1, ..., Estimated_Size_classk] |
Sample from sizes/prostate.csv :
| val_ids | val_gt_size | dumbpredwtags |
|---|---|---|
| Case00_0.nii | [147398.0, 827.0] | [140225, 6905] |
| Case00_1.nii | [147080.0, 1145.0] | [140225, 6905] |
| Case00_14.nii | [148225.0, 0.0] | [148225, 0] |
NB 1 : there should be no overlap between names of the slices in the training and validation sets (Case00_0.nii,...).
NB 2: in our implementation, the csv file contains the sizes priors in pixels, and the KL Divergence loss divides the size in pixels by (w*h) the height and weight of the slice, to obtain the class-ratio prior.
NB 3: Estimated_Size_class0 + Estimated_Size_class1 + ... + Estimated_Size_classk = w*h
NB 4: the true val_gt_size is unknown, so it is not directly used in our proposed SFDA. However, in our framework an image-level annotation is available for the target training dataset: the "Tag" of each class k, indicating the presence or absence of class k in the slice. Therefore, Estimated_Size_classk=0 if val_gt_size_k = 0 and Estimated_Size_classk>0 if val_gt_size_k > 0
NB 5: To have an idea of the capacity of the SFDA model in the ideal case where the ground truth class-ratio prior is known, it is useful to run the upper bound model SFDA_TrueSize choosing the column "val_gt_size" instead of "dumbpredwtags". This can be changed in the makefile :
results/sa/SFDA_TrueSize: OPT = --target_losses="[('EntKLProp', {'lamb_se':1,'lamb_consprior':1,'ivd':True,'weights_se':[0.1,0.9],'idc_c': [1],'curi':True,'power': 1},'PredictionBounds', \
{'margin':0,'dir':'high','idc':[0,1],'predcol':'val_gt_size','power': 1, 'mode':'percentage','sizefile':'sizes/prostate.csv'},'norm_soft_size',1)]" \
--val_target_folders="$(TT_DATA)" --l_rate 0.000001 --n_epoch 100 --lr_decay 0.9 --batch_size 10 --target_folders="$(TT_DATA)" --model_weights="$(M_WEIGHTS_ul)" \
NB 6 : If you change the name of the columns (val_ids, dumbpredwtags) in the size file, you should change them in the bounds.py file as well as in the prostate.make makefile.
results/
prostate/
fs/
best_epoch_3d/
val/
Case11_0.png
...
iter000/
val/
...
sfda/
...
params.txt # saves all the argparse parameters of the model
best_3d.pkl # best model saved
last.pkl # last epoch
IMG_target_metrics.csv # metrics over time, csv
3dbestepoch.txt # number and 3D Dice of the best epoch
...
whs/
...
archives/
$(REPO)-$(DATE)-$(HASH)-$(HOSTNAME)-sfda.tar.gz
$(REPO)-$(DATE)-$(HASH)-$(HOSTNAME)-prostate.tar.gz
The losses are defined in the losses.py file.
See below is the general structure of the target_losses parameter. The lambda weight of the loss is the last parameter.
parser.add_argument("--target_losses", type=str, required=True,
help="List of (loss_name, loss_params, bounds_name, bounds_params, fn, weight)")
Example for the supervised Cross-Entropy loss:
target_losses="[('CrossEntropy', {'idc': [0,1], 'weights':[1,1]}, None, None, None, 1)]"
Example for our SFDA loss on prostate:
target_losses="[('EntKLProp', {'lamb_se':1, 'lamb_consprior':1,'ivd':True,'weights_se':[0.1,0.9],
'idc_c': [1],'curi':True,'power': 1},'PredictionBounds', \
{'margin':0,'dir':'high','idc'[0,1],'predcol':'dumbpredwtags','power':1,
'mode':'percentage','sep':';','sizefile':'sizes/prostate.csv'},'norm_soft_size',1)]"
For the prostate and IVD applications, once you have downladed the 3D data, 2D slices can be created by the following:
python create_slices.py /path/to/IMG
python create_slices.py /path/to/GT
For the heart experiments, the data is already in 2D slices.
For the prostate experiment, as in the SAML (https://raw.githubusercontent.com/liuquande/SAML/) framework, the setting is binary segmentation, so class 1 and 2 of the original data should be merged by the following :
python remap_values.py "/path/to/source/GT" "{2:1,1:1,0:0}"
python remap_values.py "/path/to/target/GT" "{2:1,1:1,0:0}"
The new masks with will be saved in /path/to/source/GTNew and /path/to/target/GTNew.
Then, you should split the data and organize it such as in the scheme above.
In the makefiles, change the path to match the base folder containing the target dataset : T_FOLD = "your/path/to/the/target/dataset"
Run the main experiments as follows:
make -f prostate.make
make -f ivd.make
make -f whs.make
This will first run the SFDA model, which will be saved in results/sfda. It will be initialized with the networks weights obtained by training on the source only, which we made available. Eventually, you can re-run the source training model, which will be saved in results/prostate/cesource.
Remove all assertions from the code to speed up. Usually done after making sure it does not crash for one complete epoch:
make -f prostate.make <anything really> CFLAGS=-O
Use a specific python executable:
make -f prostate.make <super target> CC=/path/to/the/executable
Train for only 5 epochs, with a dummy network, and only 10 images per data loader. Useful for debugging:
make -f prostate.make <anything really> NET=Dimwit EPC=5 DEBUG=--debug
Rebuild everything even if already exist:
make -f prostate.make <a> -B
Only print the commands that will be run (useful to check recipes are properly defined):
make -f prostate.make <a> -n
This is list of parameters and hyperparameters which have influence the model's quality:
- the class-ratio anatomical prior (defined in --target_losses)
- the lambda weight for the KL loss (defined in --target_losses)
- the class weights for the KL loss (defined in --target_losses)
- the class weights for the entropy minimization loss (defined in --target_losses)
- the learning rate, and the learning rate decay
Regarding the class-ratio anatomical prior: Using the ground truth slice-based class-ratio prior (val_gt_size in the size file) is a good idea to get an idea of the capability of the SFDA on your application (it is an upper bound for our SFDA). Our main SFDA method relies on obtaining an estimation of the class-ratio for each structure of interest. Empirically, we found that an over estimation of the size of a structure yielded better quantitative and qualitative results than an under estimation^{1}. Therefore, on a new application, obtaining a coarse upper bound of an anatomical structure's pixel size should be a good starting point.
- Mathilde Bateson, Hoel Kervadec, Jose Dolz, Hervé Lombaert, Ismail Ben Ayed. Constrained Domain Adaptation for Image Segmentation. In IEEE Transactions on Medical Imaging, 2021. [paper] [implementation]
- Hoel Kervadec, Jose Dolz, Meng Tang, Eric Granger, Yuri Boykov, Ismail Ben Ayed. Constrained-CNN losses for weakly supervised segmentation. In Medical Image Analysis, 2019. [paper] [code]
- Prostate Dataset and details: https://raw.githubusercontent.com/liuquande/SAML/. The SA site dataset was used a target domain, the SB site was used as source domain. For both datasets, we use 20 scans for training, and the remaining 10 scans for validation.
- Heart Dataset and details: We used the preprocessed dataset from Dou et al. : https://github.com/carrenD/Medical-Cross-Modality-Domain-Adaptation. The data is in tfs records, it should be transformed to nii or png before running the makefile.
- Spine Dataset and details: https://ivdm3seg.weebly.com/ . From the original coronal view, we transposed the slices to transverse view in our experiments. We set the water modality (Wat) as the source and the in-phase (IP) modality as the target domain. From this dataset, 13 scans are used for training, and the remaining 3 scans for validation.
The model and code are available for non-commercial research purposes only.
{1} This could be traced to the well-known under segmentation bias of the entropy minimization alone (see our paper), which seems better compensated when the KL matching has an over segmentation bias. Note that the common DICE metric also favors oversegmentation over undersegmentation. ( See https://arxiv.org/pdf/2104.05642.pdf for example)