A Self-feedback Knowledge Elicitation Approach: Scientific Language Modeling for Chemical Reaction Predictions (SLM4CRP)
The task of chemical reaction predictions (CRPs) plays a pivotal role in advancing drug discovery and material science. However, its effectiveness is constrained by the vast and uncertain chemical reaction space and challenges in capturing reaction selectivity, particularly due to existing methods' limitations in exploiting the data's inherent knowledge. To address these challenges, we introduce a data-curated self-feedback knowledge elicitation approach. This method starts from iterative optimization of molecular representations and facilitates the extraction of knowledge on chemical reaction types (RTs). Then, we employ adaptive prompt learning to infuse the prior knowledge into the large language model (LLM). As a result, we achieve significant enhancements: a 14.2% increase in retrosynthesis prediction accuracy, a 74.2% rise in reagent prediction accuracy, and an expansion in the model's capability for handling multi-task chemical reactions. This research offers a novel paradigm for knowledge elicitation in scientific research and showcases the untapped potential of LLMs in CRPs.
Paradigm of the Review:
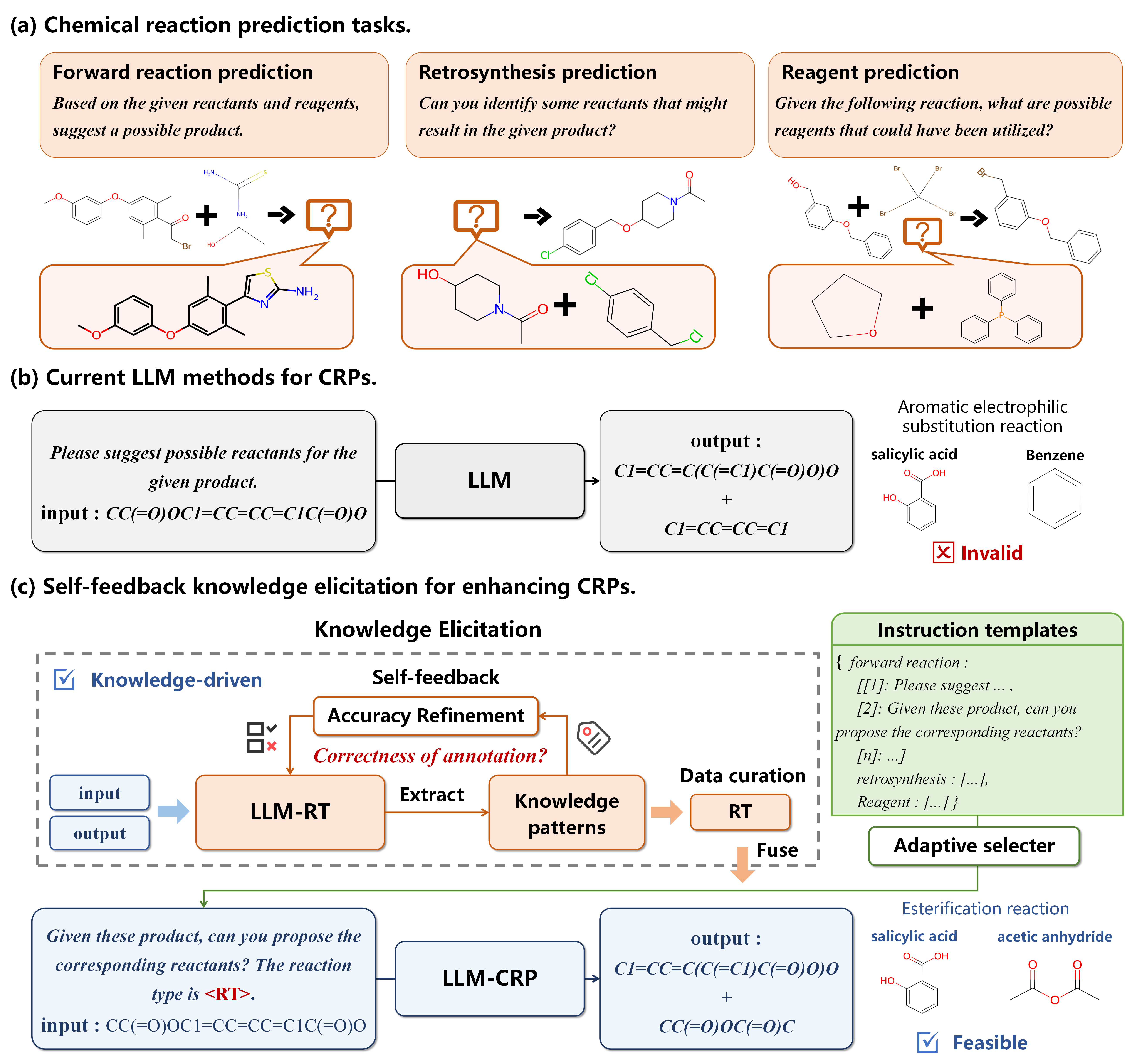
- a. Chemical reaction prediction tasks: showcasing three tasks along with examples.
- b. Current LLM methods for CRPs: indicating rational predictions but lacking in reactive validity.
- c. Self-feedback knowledge elicitation for enhancing CRPs: Self-feedback knowledge elicitation for enhancing CRPs}, demonstrating the enhancement of CRPs through the refinement of knowledge patterns, notably RTs, utilizing a self-feedback knowledge elicitation technique. Knowledge elicitation serves as a method of data curation for knowledge distillation, where RT is integrated into large language models via adaptive prompt learning, facilitating the planning of reaction pathways in CRPs.
Note: The dataset and model sections detail respective directories. Some data and checkpoints might not be available due to size constraints and permissions.
The SLM4CRP_with_RTs dataset is a CRPs dataset featuring RT labels, developed from the Mol-Instruction. We introduce a novel knowledge elicitation approach integrating a self-feedback mechanism with data curation using LLMs. This dataset embodies domain-specific knowledge by combining reactants and products of chemical reactions with annotated RTs, demonstrating that domain-integrated data can enhance the capabilities of LLMs.
forward_reaction_prediction.json: Contains data for forward reaction prediction tasks.retrosynthesis.json: Includes data for retrosynthesis tasks.reagent_prediction.json: Features data for predicting reagents in chemical reactions.reactions.json: Serves multiple tasks involving different types of chemical reactions.
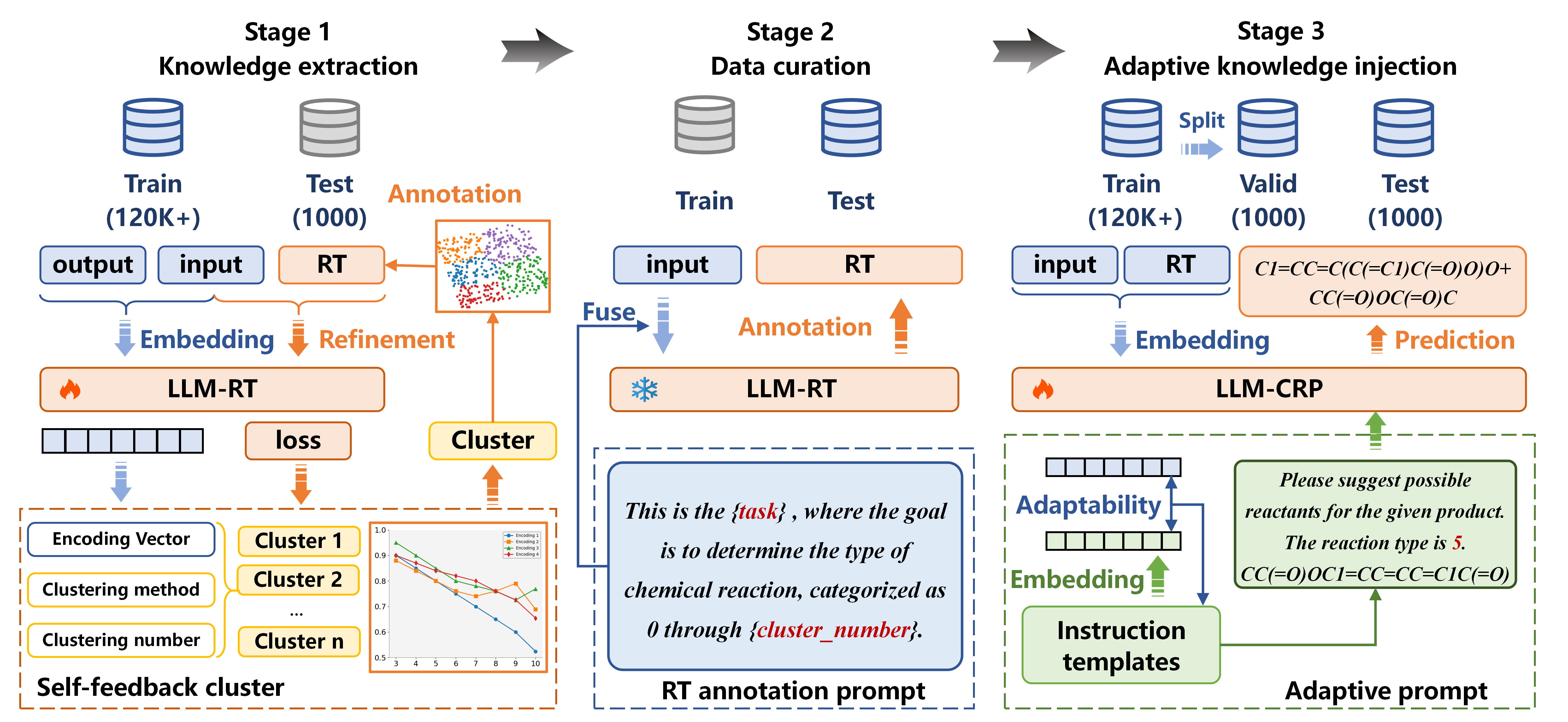
- Knowledge extraction: We divide the datasets into training, validation, and testing sets. The training set inputs and outputs are clustered using LLM-RT embeddings, facilitating RT annotations. Annotation accuracy is enhanced through iterative tuning of clustering parameters and training with inputs and RTs, aiming to pinpoint the optimal cluster settings for maximum precision.
- Data curation: The trained LLM-RT model annotates RTs for the validation and testing datasets based on their input configurations.
- Adaptive knowledge injection: Adaptability scores are derived from the embeddings of inputs and instructions, guiding the selection of the most effective adaptive instructions.
Directory containing checkpoints used for different purposes:
- finetune_ckpts: Checkpoints from fine-tuning processes.
- text_ckpts: Contains the checkpoints related to text models.
- Text+Chem T5: A model checkpoint for text and chemical data integration.
This directory includes materials for working with molecular data:
- SMILES/type: Contains training data in SMILES format.
- data_process.ipynb: Notebook for preprocessing the dataset.
Source code directory housing the implementation details:
datasets: Code for constructing datasets.dataset_manager.py: Creation of datasets for adaptive knowledge injection.dataset_manager_label.py: Creation of datasets for knowledge elicitation.
evaluations: Scripts for computing various evaluation metrics.models: Core models for the tasks.init.py: Initializes model parameters and settings.model_manager.py: Manages the loading and handling of models for knowledge injection.model_manager_label.py: Manages the loading and handling of models for knowledge elicitation.chemT5.py: Text+Chem T5.
utils: Utility functions and initializations.init.py: General utility tool initialization.xutils.py: Advanced and specialized utility tool initialization.
task_manager.py: Function to execute tasks related to adaptive knowledge injection.task_manager_label.py: Function to execute tasks related to knowledge elicitation.
mode: Select the operation mode. Options includedata_check,encoder_check,train, andeval.N: Number of clusters.reaction_type: Specifies whether to include RT during training.task: Task type. Options includeforward,retro,reagent, andreactions.batch_size: Set the batch size for operations.
-
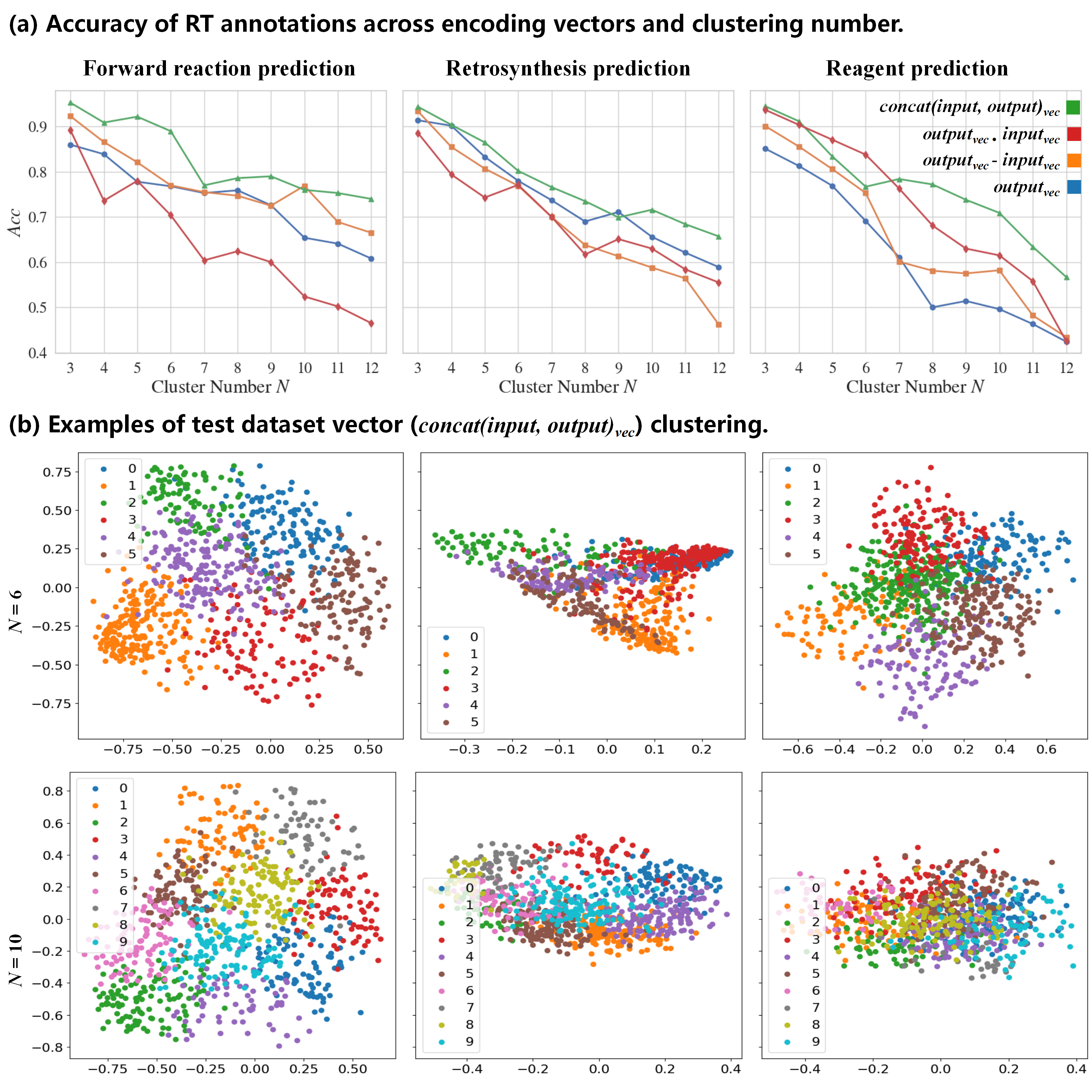
Accuracy of RT annotations across encoding vectors and clustering numbers: We assess the annotation accuracy (
$Acc$ ) among four encoding methods at various reasonable clustering numbers ($N$ ). The results reveal that the encoding method utilizing concatenated input-output vectors ($concat(input, output)_{vec}$ ) delivers the highest performance. -
Clustering of test dataset vectors (
$concat(input, output)_{vec}$ ): With$N$ set to 6 and 10, test dataset vectors are processed through a linear layer to reduce them to two dimensions, effectively visualizing the clustering outcomes.
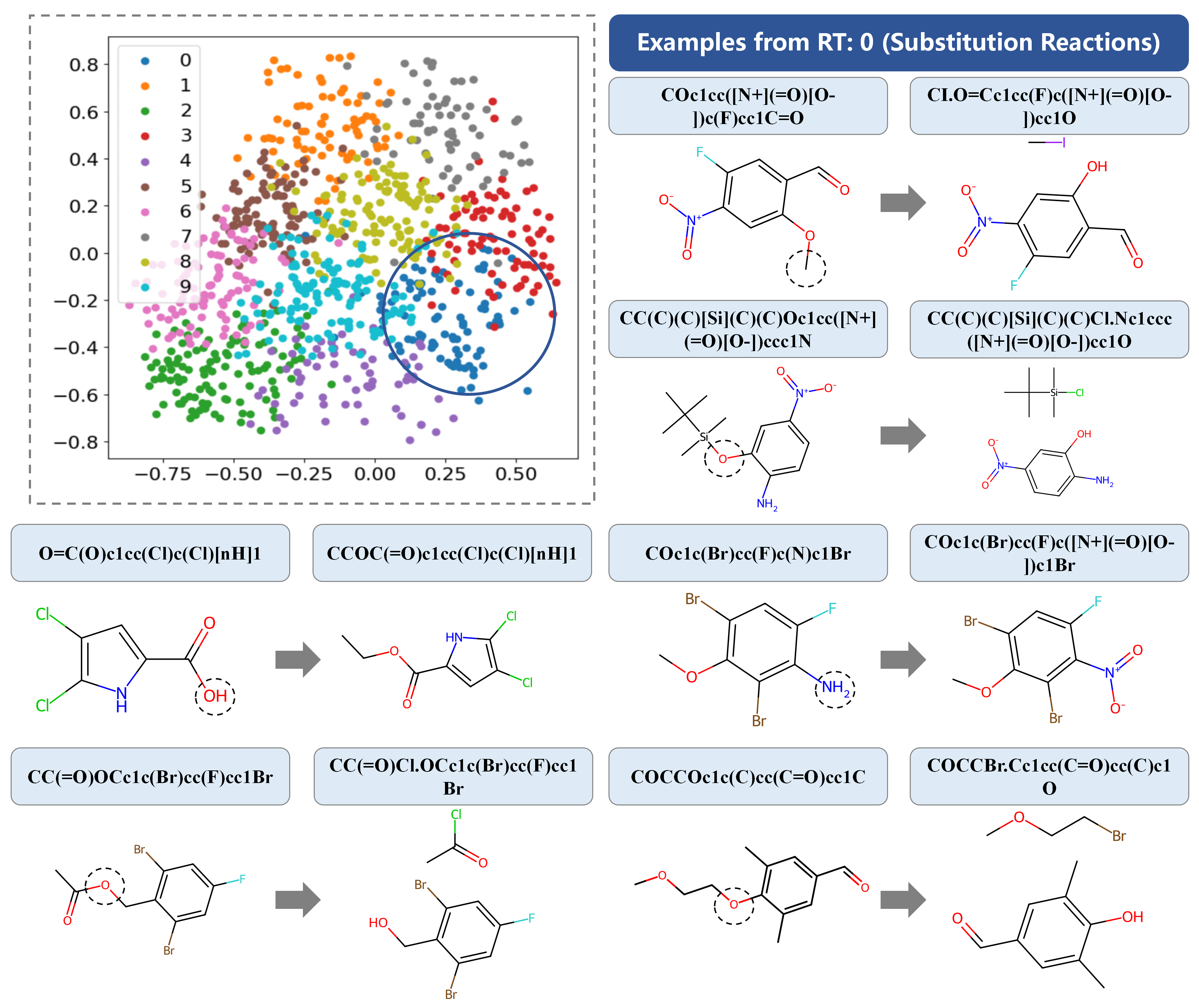
To validate the practical significance of RT annotation, we analyze samples filtered through the concat(input, output)_{vec} vector with N=10 labeled results, focusing on samples with an RT label of 0. These instances typically involve simple atomic substitutions, verifying the predominance of substitution reactions in these cases. This analysis highlights the real-world relevance of our RT annotation method.
@misc{liu2024selffeedback,
title={A Self-feedback Knowledge Elicitation Approach for Chemical Reaction Predictions},
author={Pengfei Liu and Jun Tao and Zhixiang Ren},
year={2024},
eprint={2404.09606},
archivePrefix={arXiv},
primaryClass={cs.LG}
}
The development of the SLM4CRP_with_RTs dataset was greatly inspired by the Mol-Instruction approach to CRPs. We are also thankful to Hugging Face for providing the initial model weights that facilitated our research.