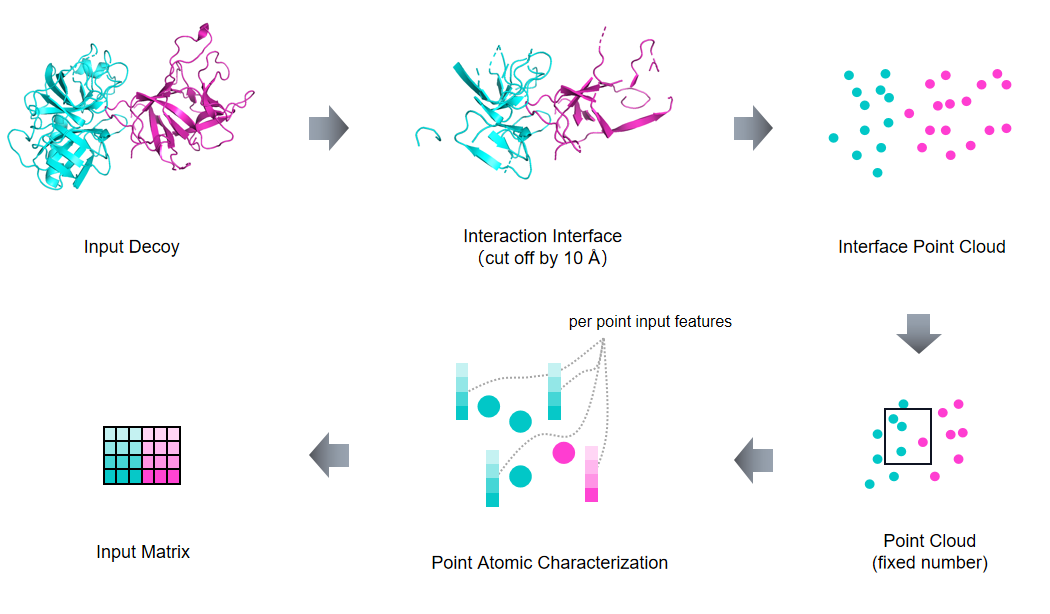
PointDE is a 3D point cloud neural network for protein docking evaluation.
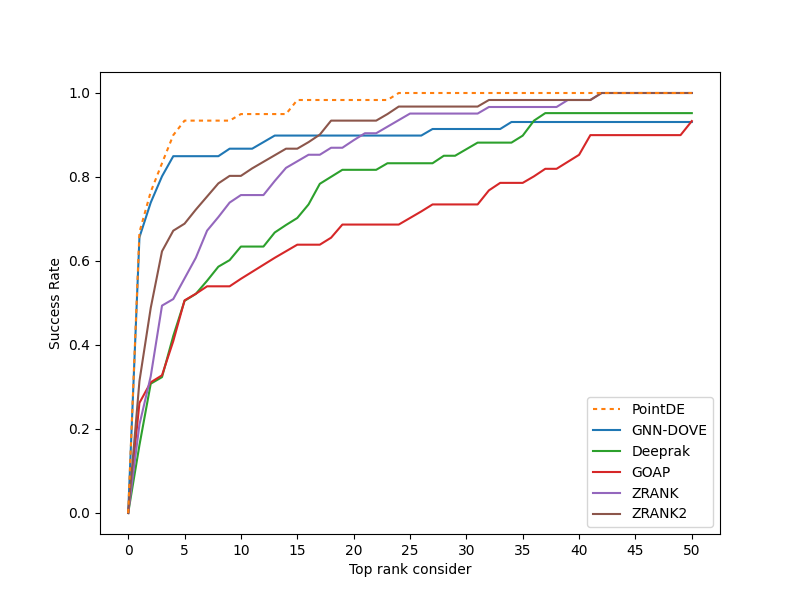
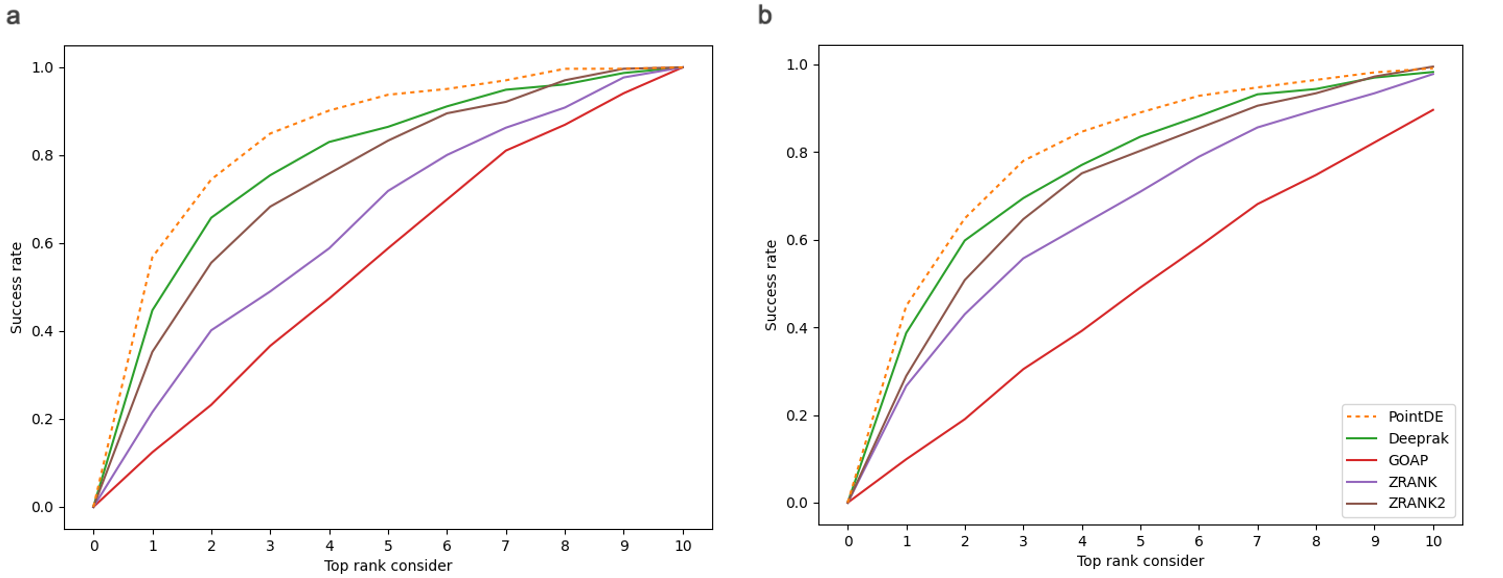
Protein-protein interactions (PPIs) play essential roles in many vital movements and The determination of protein complex structure is helpful to discover the mechanism of PPI. Protein-protein docking is being developed to modeling the structure of protein. However, there is still a challenge to select the near-native decoys generated by protein-protein docking. Here, we propose a docking evaluation method using 3D point cloud neural network named PointDE. PointDE transform protein structure to point cloud. Using the state-of-the-art point cloud network architecture and a novel grouping mechanism, PointDE can capture the geometries of point cloud and learn the interaction information from protein interface. On public datasets, PointDE surpasses the state-of-the-art method using deep learning. To further explore the ability of our method in different type protein structures, we developed a new dataset generated by high-quality antibody-antigen complexes. The result in this antibody-antigen dataset shows the strong performance of PointDE, which will be helpful for the understanding of PPI mechanisms.
- Python 3.6
- Torch 1.5.1
- scikit-learn
- torchsampler
1. Install Git
git clone https://github.com/AI-ProteinGroup/PointDE && cd PointDE
conda create -n PointDE python=3.6
conda activate PointDE
pip install -r requirements.txt
That downloads the models for cross-validation on DOCKGROUND.
python preprocess.py --p_num [process_number] --npoint [N] --dataset_dir [dataset_dir] --data_sv_dir [special_data_dir]
preprocessing should specify a pdb file with Receptor chain ID 'A' and ligand chain ID 'B'. pdb file must be in a folder named PDB ID. num_workers is used to specify the number of theads to process data.
File Example:
dataset dir
│
└───1A2Y
│ │ 1A2Y_01.pdb
│ │ 1A2Y_02.pdb
│ │ ...
│
└───1A2K
│ │ 1A2K_01.pdb
│ │ 1A2K_02.pdb
│ │ ...
│
python train.py --data_dir [data_dir] --gpu=[gpu_id] --batch_size [batch_size] --checkpoint [checkpoint_dir]
After preprocessing the data, you are responsible for generating the division of training and testing data in the format specified in "document/training_annotation_example". Please ensure that you place these files in the preprocessed dataset folder.
main.py should specify a file preprocessed by preprocess.py; "gpu_id" is used to specify the gpu id; trianing model will be saved in "checkpoint_dir". "data_dir" is the path saved in the previous step "data_sv_dir".
The models for cross-validation on DOCKGROUND here.
python eval.py --gpu=[gpu_id] --fold [fold] --data_dir [data_dir] --sv_dir [sv_dir]
eval.py should specify the directory that inclues pdb files with Receptor chain ID 'A' and ligand chain ID 'B'; --fold should specify the fold model you will use, where -1 denotes that you want to use the average prediction of 4 fold models and 1,2,3,4 will choose different model for predictions. The output will be kept in [ssr_sv/{sv_dir}]. The prediction results will be kept in {PDB ID}.txt.