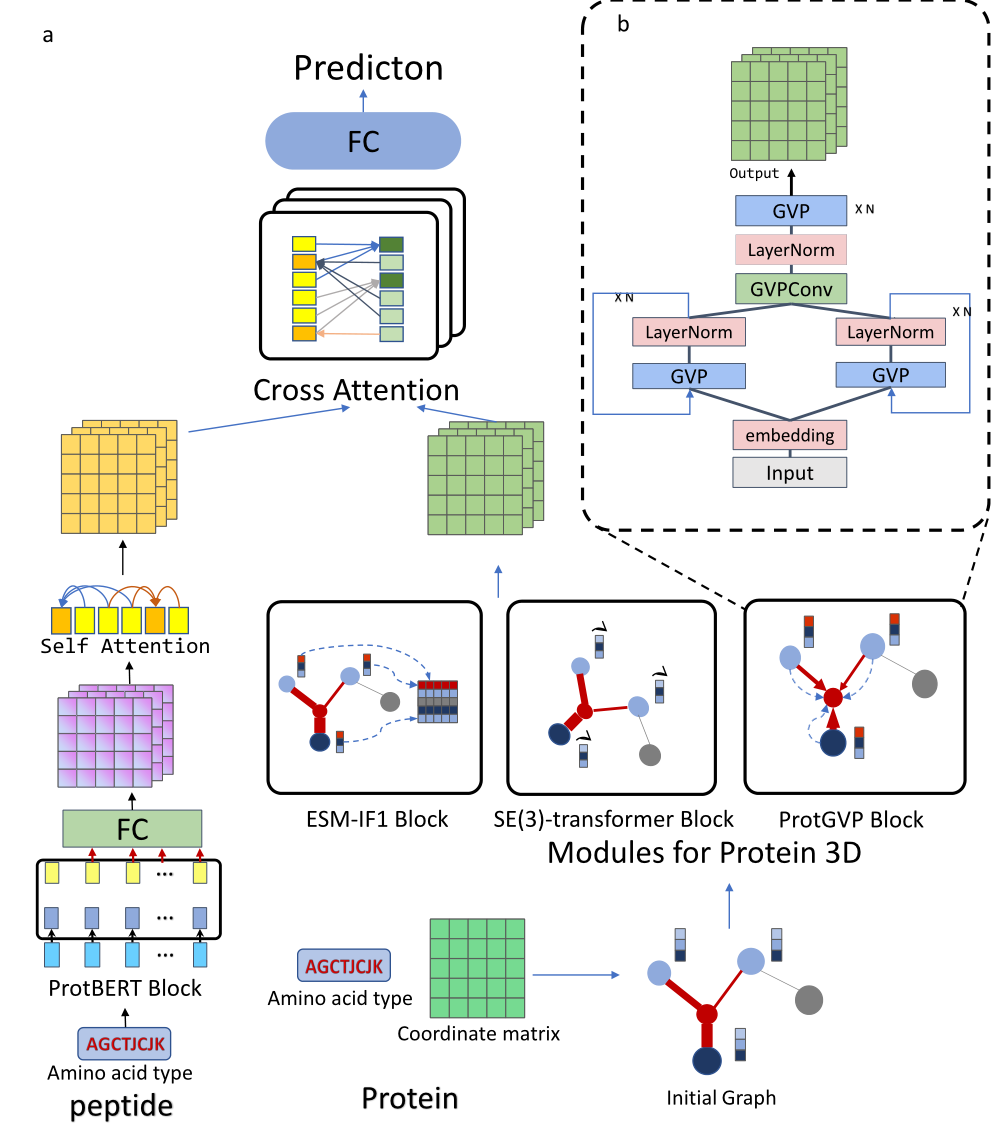
We think the requirements for the environment are so loose that any version can be used
cuda 10.2
conda install gxx_linux-64 7.3
conda install pytorch==1.8.1 torchvision==0.9.1 torchaudio==0.8.1 cudatoolkit=10.2 -c pytorch
conda install pyg -c pyg -c conda-forge
conda install tqdm
conda install -c jrybka sklearn
conda install -c conda-forge tensorboard
conda install -c conda-forge jsonlines
pepbdb data:http://huanglab.phys.hust.edu.cn/pepbdb/ propedia data:http://bioinfo.dcc.ufmg.br/propedia/download
Detailed data processing steps are given in the readme of the dataprocess
Go to the ESM-IF1 folder and run the train_mySE3.py file
python train_esm.pyEmpty logs/, models/, and save/ folders is needed to store procedures, breakpoints, and final results.
Go to the SE(3)-Tansformer folder and run the train_mySE3.py file
python train_mySE3.pyEmpty logs/, models/, and save/ folders is needed to store procedures, breakpoints, and final results.
Go to the ProtGVP folder and run the train_ProtGVP.py file
python train_ProtGVP.pyEmpty logs/, models/, and save/ folders is needed to store procedures, breakpoints, and final results.
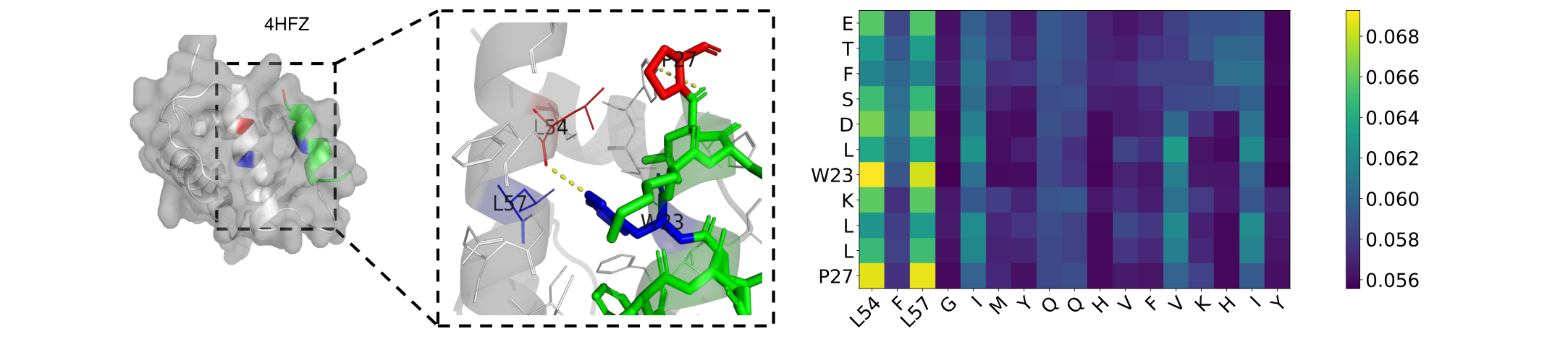
Fig. 4 Visualization and Attention matrix on 4HFZ. L54 forms a hydrogen bond with W23 (yellow line segment), where L57 is physically close to the W53 amino acid (blue) and P27 is physically close to L54 (red).
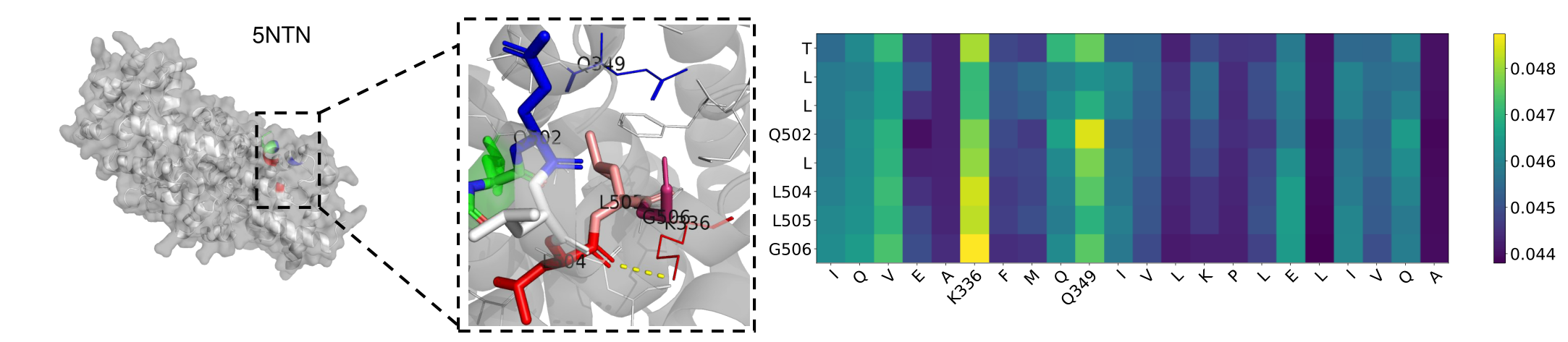
Fig. 5 Visualization and Attention matrix on 5NTN. L504 forms a hydrogen bond with K336 (yellow line segment), where L505 is physically close to K336 amino acid (pink), G506 is physically close to K336 (Burgundy), and Q502 is physically close to Q349 (blue).
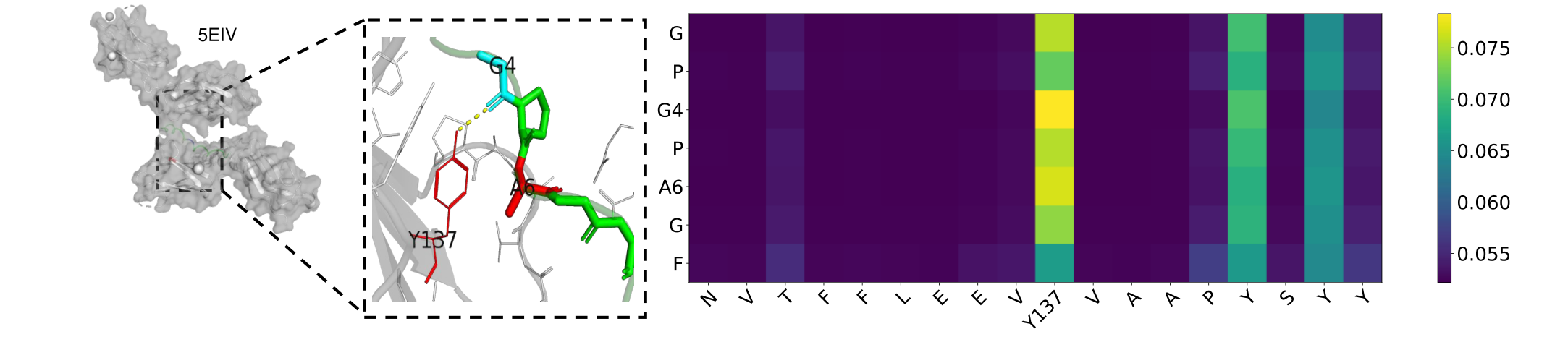
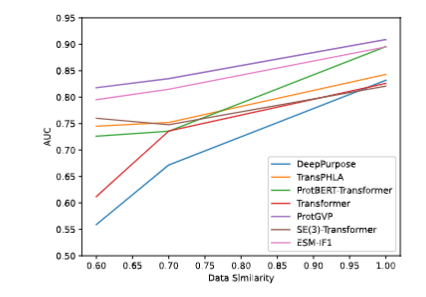
Fig. 6 Visualization and Attention matrix on 5EIV. G4 creates a hydrogen bond with Y137 (yellow line segment), where A6 is physically close to the amino acid Y137 (red)Fig.2 Changes in AUC under Different Similarities of Each Model.
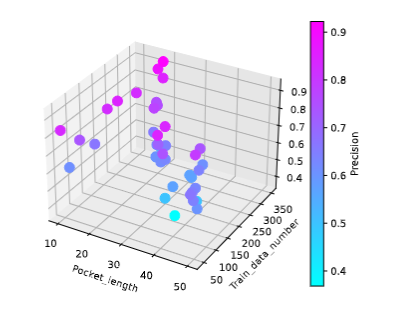
Fig.4 The relationship between precision and the amount of training data and pocket size.
More details will be given in the future.