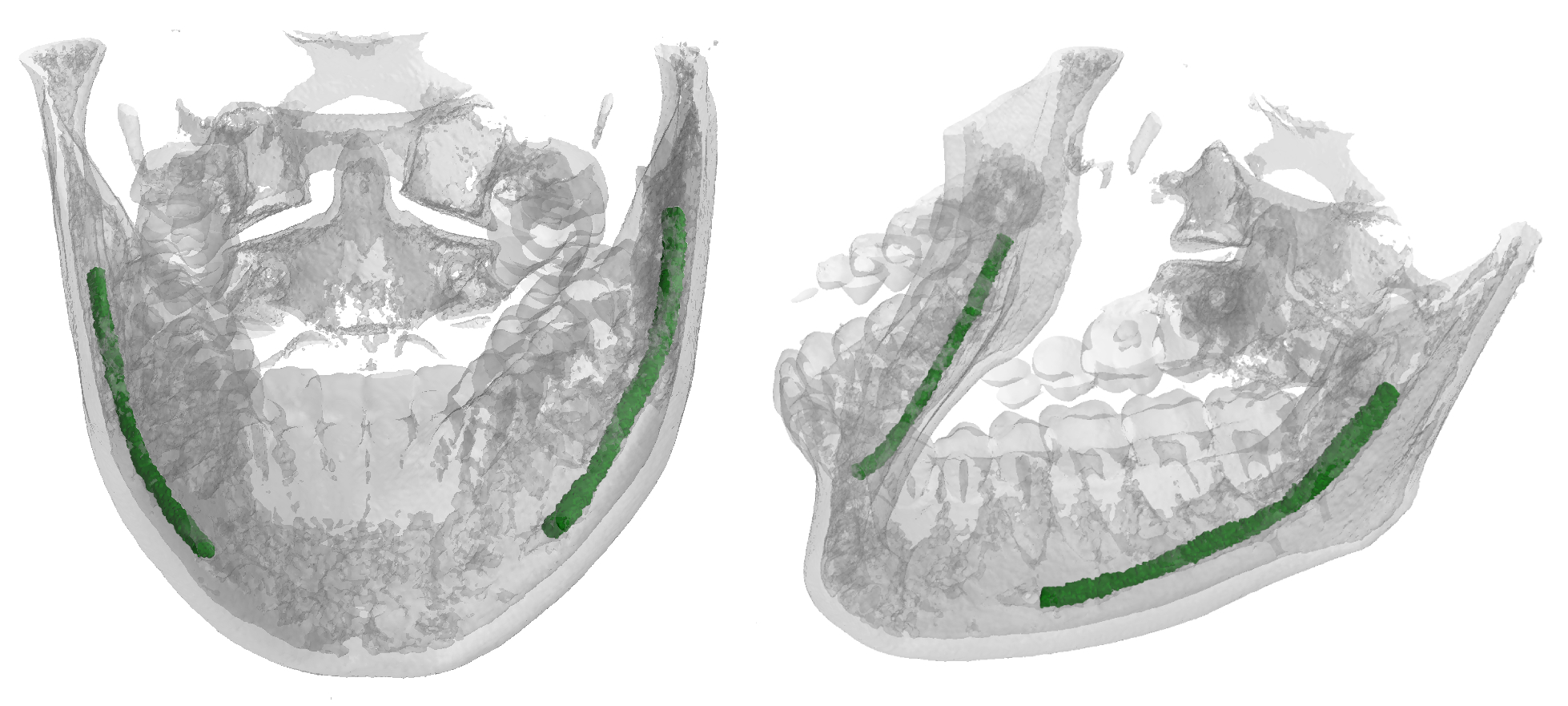
This repository contains the material from the paper "Improving Segmentation of the Inferior Alveolar Nerve through Deep Label Propagation". In particular, this repo is dedicated to the 3D neural networks used to generate and segment the Inferior Alveolar Nerve (IAN). This nerve is oftentimes in close relation to the roots of molars, and its position must thus be carefully detailed before the surgical removal. As avoiding contact with the IAN is a primary concern during these operations, segmentation plays a key role in surgical preparations.
For the IAN segmentation, we adopted a modified version of U-NET 3D, enriched with a 2 pixels padding and an embedding of the coordinates of the sub-volumes fed in the net. Because of the heavy burden represented by manual annotation of segmentation ground thruth we also employed the same neural network to expand our dataset by having a dense annotation for all the volumes in our dataset in order to have more data for the training phase of the main task. The training phase of the segmentation network, is divided in 2 phase: first we use the sparse annotations with their generated ground thruth as a pretraining and then the real dense annotation as a finetuning.
Before running this project, you need to download the dataset. Also look at this which has the code to generate the naive dense labels starting from the sparse annotations (Circular Expansion)
Clone this repository, create a python env for the project (optional) and activate it. Then install all the dependencies with pip
git clone git@github.com:AImageLab-zip/alveolar_canal.git
cd alveolar_canal
python -m venv env
source env/bin/activate
pip install -r requirements.txt
Run the project as follows:
python main.py [-h] -c CONFIG [--verbose]
arguments:
-h, --help show this help message and exit
-c CONFIG, --config CONFIG
the config file used to run the experiment
--verbose To log also to stdout
E.g. to run the generation experiment, execute:
python main.py --config configs/gen-training.yaml
You can find the config files used to obtain the best result in the config folder.
Two files are needed: experiment.yaml, augmentations.yaml. For both the two
tasks, the best config file is provided:
- gen-training.yaml for the network which, from the sparse annotation, generate the dense labels
- seg-pretrain.yaml which train the segmentation network only over the generated labels
- seg-finetuning.yaml which train the segmentation network over the real dense labels
Execute
main.pywith these 3 configs in this order to reproduce our results
Download the pre-trained checkpoints here
experiment.yaml describe each part of the project, like the
network/loss/optimizer, how to load data and so on:
# title of the experiment
title: canal_generator_train
# Where to output everything, in this path a folder with
# the same name as the title is created containing checkpoints,
# logs and a copy of the config used
project_dir: '/path/to/results'
seed: 47
# which experiment to execute: Segmentation or Generation
experiment:
name: Generation
data_loader:
dataset: /path/to/maxillo
# null to use training_set, generated to used the generated dataset
training_set: null
# which preprocessing to use, see: preprocessing.yaml
preprocessing: configs/preprocessing.yaml
# which augmentations to use, see: augmentations.yaml
augmentations: configs/augmentations.yaml
background_suppression: 0
batch_size: 2
labels:
BACKGROUND: 0
INSIDE: 1
mean: 0.08435
num_workers: 8
# shape of a single patch
patch_shape:
- 120
- 120
- 120
# reshape of the whole volume before extracting the patches
resize_shape:
- 168
- 280
- 360
sampler_type: grid
grid_overlap: 0
std: 0.17885
volumes_max: 2100
volumes_min: 0
weights:
- 0.000703
- 0.999
# which network to use
model:
name: PosPadUNet3D
loss:
name: Jaccard
lr_scheduler:
name: Plateau
optimizer:
learning_rate: 0.1
name: SGD
trainer:
# Reload the last checkpoints?
reload: True
checkpoint: /path/to/checkpoints/last.pth
# train the network
do_train: True
# do a single test of the network with the loaded checkpoints
do_test: False
# generate the synthetic dense dataset
do_inference: False
epochs: 100preprocessing.yaml defines which type of preprocessing to use during training.
One simple preprocessing file has been used for every experiment.
The file should follow this structure:
Clamp:
out_min: 0
out_max: 2100
RescaleIntensity:
out_min_max: !!python/tuple [0, 1]augmentations.yaml defines which type of augmentations use during training.
Two different augmentations files have been used, one for the segmentation task,
one for the generation task.
The file should follow this structure:
RandomAffine:
scales: !!python/tuple [0.5, 1.5]
degrees: !!python/tuple [10, 10]
isotropic: false
image_interpolation: linear
p: 0.5
RandomFlip:
axes: 2
flip_probability: 0.7