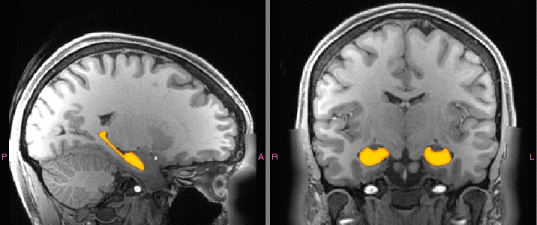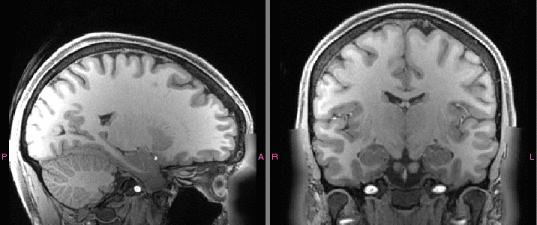
Brain Hippocampus Segmentation
This program can quickly segment (<2min) the Hippocampus of raw brain T1 images.
It relies on a Convolutional Neural Network pre-trained on thousands of images from multiple large cohorts, and is therefore quite robust to subject- and MR-contrast variation. For more details on how it has been created, refer to the corresponding manuscript at http://dx.doi.org/10.1016/j.media.2017.11.004
A PyTorch version of this program is available at https://github.com/bthyreau/hippodeep_pytorch. It is now recommended to use it instead.
(new) (Apr 2018) improved the initial registration step, for a low speed penalty
This program requires Theano >= 0.9.0 and Lasagne
No GPU is required
The ANTs tools are required for the initial low-res registration. Optionally, the FSL tools can be used as alternative.
Tested on Linux CentOS 6.x and 7.x, and MacOS X
The code uses the Scipy stack and a recent Theano. It also requires the nibabel library (for nifti loading) and the Lasagne library.
To setup a ANTs environment, get it from http://stnava.github.io/ANTs/ (or alternatively, from a docker container such as http://www.mindboggle.info/ )
The simplest way to install the rest from scratch is to use a Anaconda environment, then
- install scipy and Theano >=0.9.0 (
conda install theano) - nibabel is available on pip (
pip install nibabel) - Lasagne (version >=0.2 If still not available, it should be probably pulled from the github repo
pip install --upgrade https://github.com/Lasagne/Lasagne/archive/master.zip)
After download, you can run
./first_run.sh in the source directory to ensure the environment is ok and to pre-compile the models.
Then, to use the program, simply run:
deepseg1.sh example_brain_t1.nii.gz.
The resulting segmentation should be stored as example_brain_t1_mask_L.nii.gz (or R for right).
The volumes values are stored in example_brain_t1_hippo_vol_LR.txt, Left then Right, in mm^3. Also, example_brain_t1_eTIV.txt has an estimate of the Intracranial-Volume.
(hint) to concatenate multiple outputs into a table, you can use a bash shell such as for a in *_hippo_vol_LR.txt ; do cat $a ${a/hippo_vol_LR/eTIV} | xargs echo $a; done
Alternatively, if you wish to re-use the same version as described in the manuscript, then you can either run:
Using ANTs tools: deepseg2.sh example_brain_t1.nii.gz.
or using FSL tools: deepseg3.sh example_brain_t1.nii.gz,
The script can be called with its full path, but the image file must be in current, writable, directory.