Lars Mølgaard Saxhaug 6/25/2021
library(tidyverse)
library(brms)
library(tidybayes)
library(here)
theme_set(theme_tidybayes())regenron <- tribble(
~"group", ~"intervention", ~"death", ~"n",
"seropos", "regenron", 411, 2636,
"seropos", "control", 383, 2636,
"seroneg", "regenron", 396, 1633,
"seroneg", "control", 451, 1520,
"unknown", "regenron", 137, 570,
"unknown", "control", 192, 790
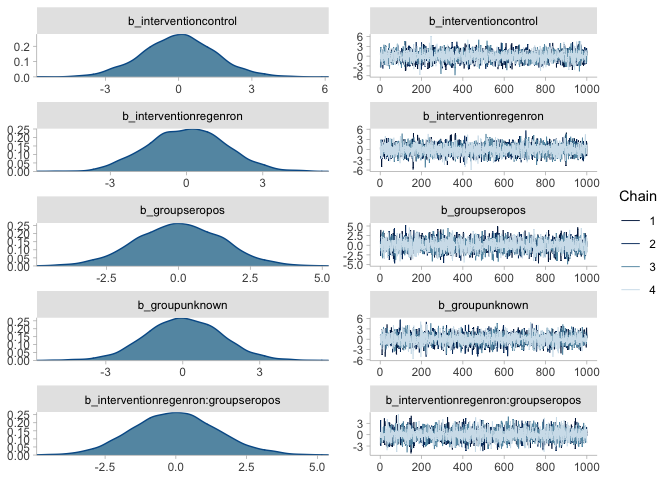
) %>% mutate(across(where(is.character), as.factor))mr_prior <- brm(death | trials(n) ~ 0 + intervention * group, # formula without intercept
family = binomial(), data = regenron,
prior = prior(normal(0, 1.5), class = "b"),
sample_prior = "only",
file = here("model_fits", "mr_prior"),
file_refit = "on_change"
)
summary(mr_prior)## Family: binomial
## Links: mu = logit
## Formula: death | trials(n) ~ 0 + intervention * group
## Data: regenron (Number of observations: 6)
## Samples: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
## total post-warmup samples = 4000
##
## Population-Level Effects:
## Estimate Est.Error l-95% CI u-95% CI Rhat
## interventioncontrol 0.01 1.49 -2.92 2.98 1.00
## interventionregenron 0.01 1.53 -2.87 2.99 1.00
## groupseropos 0.03 1.49 -2.92 2.94 1.00
## groupunknown 0.04 1.48 -2.88 2.95 1.00
## interventionregenron:groupseropos -0.02 1.49 -2.90 3.06 1.00
## interventionregenron:groupunknown 0.01 1.47 -2.93 2.87 1.00
## Bulk_ESS Tail_ESS
## interventioncontrol 5281 3106
## interventionregenron 5736 2904
## groupseropos 5755 2798
## groupunknown 5663 3251
## interventionregenron:groupseropos 5337 2688
## interventionregenron:groupunknown 5554 2736
##
## Samples were drawn using sampling(NUTS). For each parameter, Bulk_ESS
## and Tail_ESS are effective sample size measures, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).
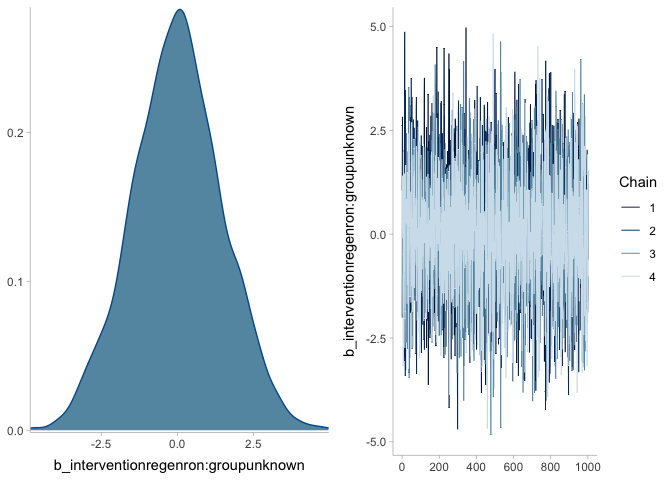
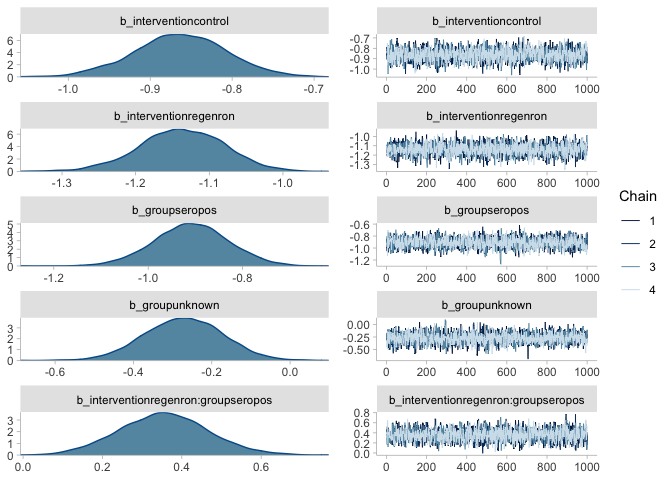
plot(mr_prior)mr <- brm(death | trials(n) ~ 0 + intervention * group,
family = binomial(),
data = regenron,
prior = prior(normal(0, 1.5), class = "b"),
file = here("model_fits", "mr"),
file_refit = "on_change"
)
summary(mr)## Family: binomial
## Links: mu = logit
## Formula: death | trials(n) ~ 0 + intervention * group
## Data: regenron (Number of observations: 6)
## Samples: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
## total post-warmup samples = 4000
##
## Population-Level Effects:
## Estimate Est.Error l-95% CI u-95% CI Rhat
## interventioncontrol -0.86 0.06 -0.98 -0.75 1.00
## interventionregenron -1.14 0.06 -1.26 -1.03 1.00
## groupseropos -0.91 0.08 -1.07 -0.76 1.00
## groupunknown -0.27 0.10 -0.47 -0.08 1.00
## interventionregenron:groupseropos 0.36 0.11 0.14 0.58 1.00
## interventionregenron:groupunknown 0.25 0.15 -0.04 0.55 1.00
## Bulk_ESS Tail_ESS
## interventioncontrol 1980 2457
## interventionregenron 2800 2430
## groupseropos 2049 2583
## groupunknown 2226 2449
## interventionregenron:groupseropos 2062 2423
## interventionregenron:groupunknown 2200 2398
##
## Samples were drawn using sampling(NUTS). For each parameter, Bulk_ESS
## and Tail_ESS are effective sample size measures, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).
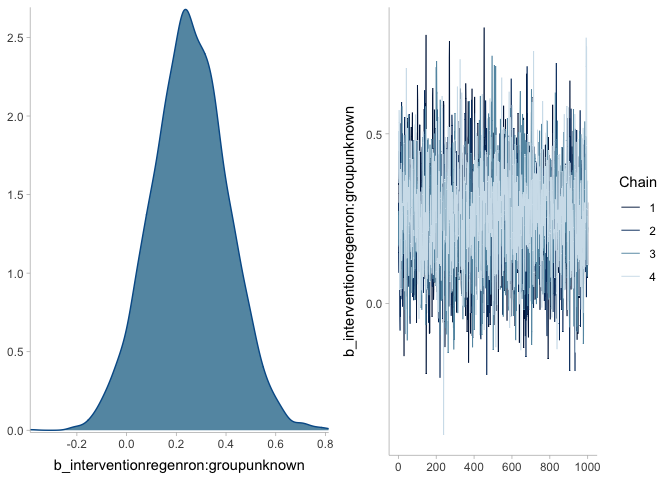
plot(mr, ask = FALSE)regenron %>% # original data
modelr::data_grid(group, intervention) %>% # generate new data
mutate(n = 1) %>% # n is the number of trials
add_fitted_draws(mr) %>% # compute draws from the linear predictor for model `mr`, replace with `mr_prior` for prior prediction
mutate(.value = brms::inv_logit_scaled(.value)) %>% # inverse logit transformation to probability scale
compare_levels(.value, by = intervention, fun = `-`) %>% # calculate absolute risk reduction per group
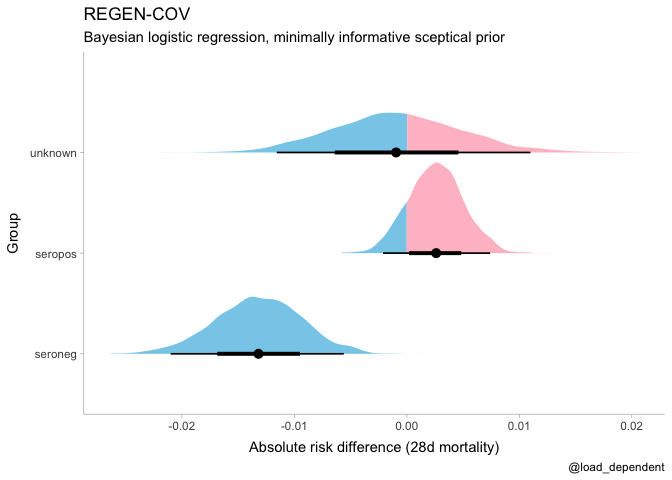
ggplot(aes(x = .value, y = group, fill = after_stat(ifelse(x > 0, "over", "under")))) +
stat_halfeye() +
scale_fill_manual(values = c("over" = "pink", "under" = "skyblue")) +
theme(legend.position = "none") +
scale_y_discrete(name = "Group") +
scale_x_continuous(name = "Absolute risk difference (28d mortality)") +
labs(title = "REGEN-COV", subtitle = "Bayesian logistic regression, minimally informative sceptical prior", caption = "@load_dependent")regenron %>%
modelr::data_grid(group, intervention) %>%
mutate(n = 1) %>%
add_fitted_draws(mr) %>%
compare_levels(.value, by = intervention) %>%
mutate(or = exp(.value)) %>%
group_by(group) %>%
summarise(`Probability of superiority` = mean(or < 1)) %>%
mutate(group=str_to_title(group)) %>%
knitr::kable(col.names = c("Group","Probability of superiority"))| Group | Probability of superiority |
|---|---|
| Seroneg | 0.99975 |
| Seropos | 0.15125 |
| Unknown | 0.56750 |