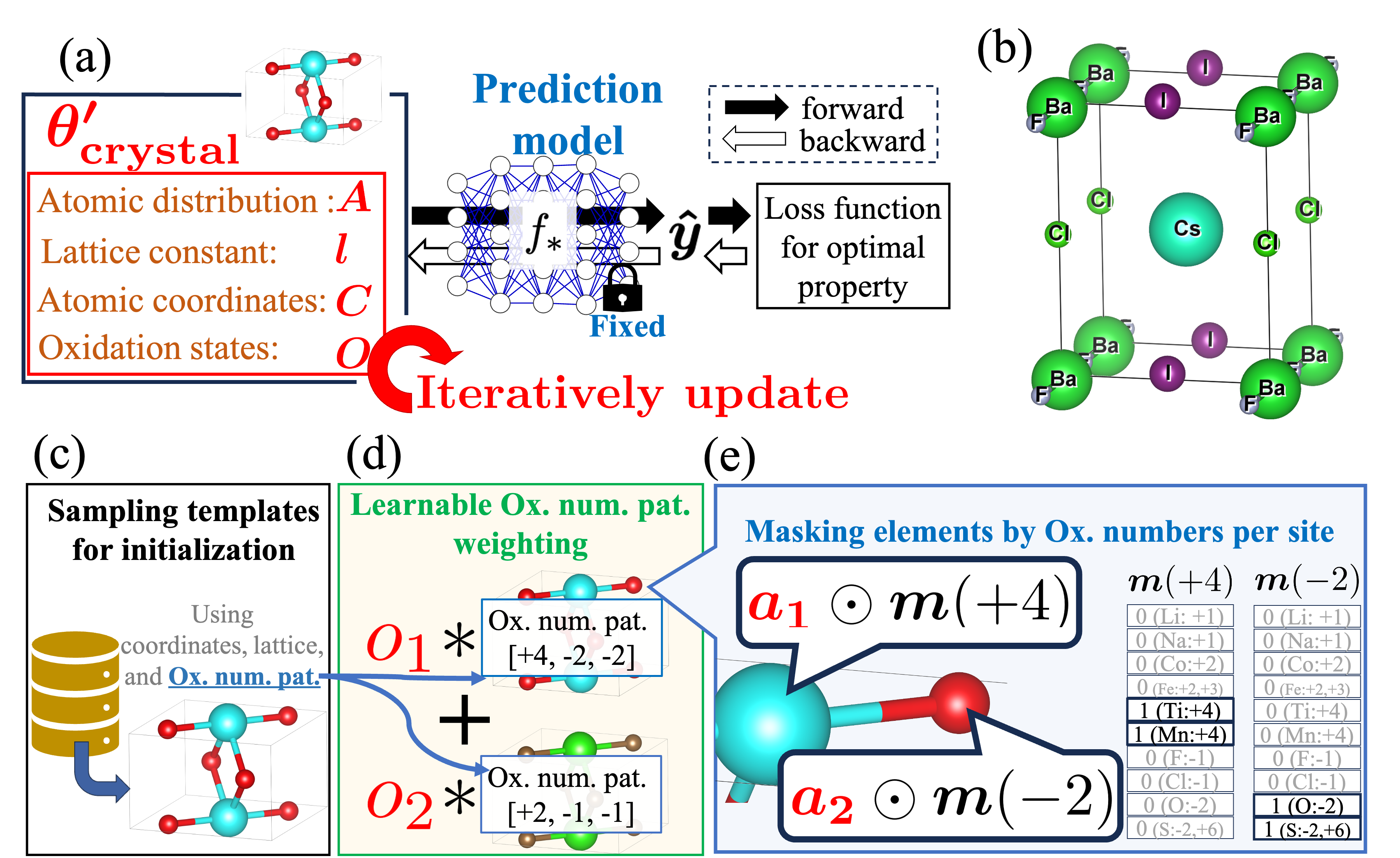
SMOACS is a framework that utilizes state-of-the-art property prediction models and their gradients to directly optimize input crystal structures for multiple targeted properties simultaneously. It enables the integration of adaptive constraints into the optimization process without necessitating model retraining. This allows for optimizing targeted properties while maintaining specific crystal structures, such as perovskites, even with models trained on diverse crystal types.
For detailed information, please refer to our paper:
If you use this work in your research, please cite:
@article{afujii2024,
title={Adaptive Constraint Integration for Simultaneously Optimizing Crystal Structures with Multiple Targeted Properties},
author={Akihiro FUJII, Yoshitaka Ushiku, Koji Shimizu, Anh Khoa Augustin Lu, Satoshi Watanabe },
journal={arXiv preprint arXiv:2410.08562},
year={2024}
}
The code in this repository uses the code of Crystalformer and ALIGNN. Crystalformer's pretrained models provided in Crystalformer's official repository and ALIGNN's pretrained models provided in ALIGNN's official repository. Thus, if you use our code in your projects, please cite both our work and the Crystalformer paper or ALIGNN:
@inproceedings{taniai2024crystalformer,
title = {Crystalformer: Infinitely Connected Attention for Periodic Structure Encoding},
author = {Tatsunori Taniai and
Ryo Igarashi and
Yuta Suzuki and
Naoya Chiba and
Kotaro Saito and
Yoshitaka Ushiku and
Kanta Ono
},
booktitle = {The Twelfth International Conference on Learning Representations},
year = {2024},
url = {https://openreview.net/forum?id=fxQiecl9HB}
}
@article{choudhary2021atomistic,
title={Atomistic line graph neural network for improved materials property predictions},
author={Choudhary, Kamal and DeCost, Brian},
journal={npj Computational Materials},
volume={7},
number={1},
pages={185},
year={2021},
publisher={Nature Publishing Group UK London}
}
torch==2.2.1
pymatgen==2023.10.11
SMACT==2.2.5
jarvis-tools==2023.9.20
dgl==1.1.2
alignn==2024.4.10
numpy==1.25.2
You need to download the pretrained weights for Crystalformers from their GitHub page and place them in the models/crystalformer directory. The weights for ALIGNN are automatically downloaded from figshare.
Run python main.py for a small demonstration. Modify the arguments as necessary.
If you want to use larger size of initial crystal structures dataset, please run python create_data.py. You can find larger size of initial crystal structures dataset in ./data.
Run python main.py for a demonstration. Modify the arguments as needed.
If you'd like to use a larger initial crystal structure dataset, please run 'python create_data.py'. You can find the larger dataset in the './data' directory.
main_experiment(
dir_path = './initial_crystal_candidates/structures_from_MEGNet', # directory of initial crystal structures
mask_data_path = './mask_arrays/mask_dict_super_common.npz', # atomic mask array
radii_data_path = None, # ionic radii array
exp_name = 'S_Cry_random', # experiment name.
model_name = 'Crystalformer' , # model name. 'Crystalformer' or 'ALIGNN'
num_steps=200, # optimization steps
num_candidate=8, # The number of initial crystal structures
num_batch_crystal=8, # Batch size to optimize
# prediction model, loss and criteria for each target values
prediction_loss_setting_dict ={
'bandgap': {
'prediction_model':None,
'loss_function':{
'func_name':'GapLoss',
'margin':0.04, # (eV)
'target_bandgap':2.0, # (eV)
'mode':'band',
'coef':1.0, # coefficient for the loss
},
'criteria_max':2.04, # optimized values should be less than this value
'criteria_min':1.96, # optimizedvalues should be more than this value
},
'e_form':{
'prediction_model':None,
'loss_function':{
'func_name':'FormationEnegryLoss',
'e_form_min':None,
'e_form_coef':1.0, # coefficient for the loss
},
'criteria_max':-0.5, # optimized values should be less than this value
'criteria_min':None, # optimizedvalues should be more than this value
}
},
atomic_dictribution_loss_setting_dict = {},
site_atom_oxidation_dict = {'Atom_settings': {}, 'use_ionic_radii':False},
limit_coords_displacement= None, # limiation for displacement of fractional coordindates
lattice_lr= 0.01, # learning rate for lattice (a,b,c,α,β,γ) optimization
coords_lr= 0.02, # learning rate for fractional coordinates
atom_lr= 0.6, # learning rate for atomic distribution and oxidation state configuration parameters
) | file path | description |
|---|---|
.results/{exp_name}/result_summary.csv |
summary scores |
.results/{exp_name}/result.csv |
scores for each optimized crystal structures |
.results/{exp_name}/result.npz |
detailed scores with numpy |
.results/{exp_name}/poscar/*.vasp |
optimized crystal structures written as POSCAR files |
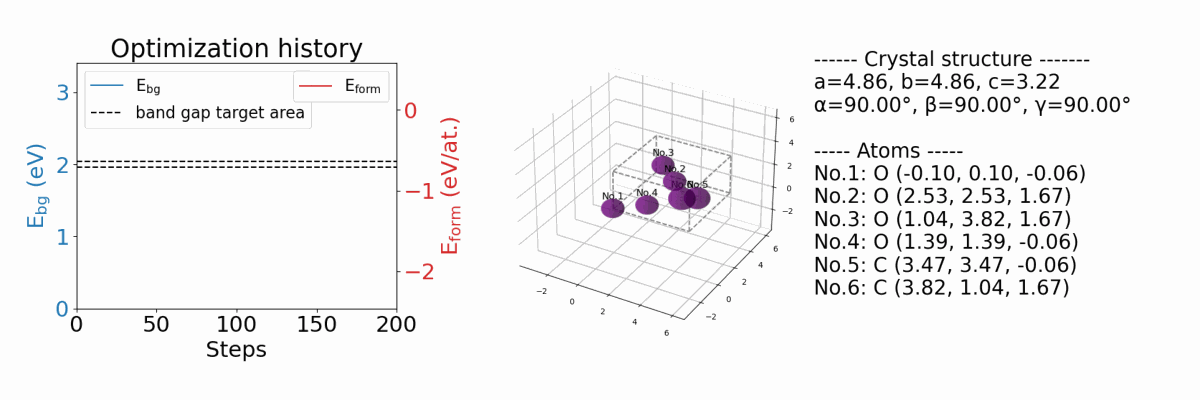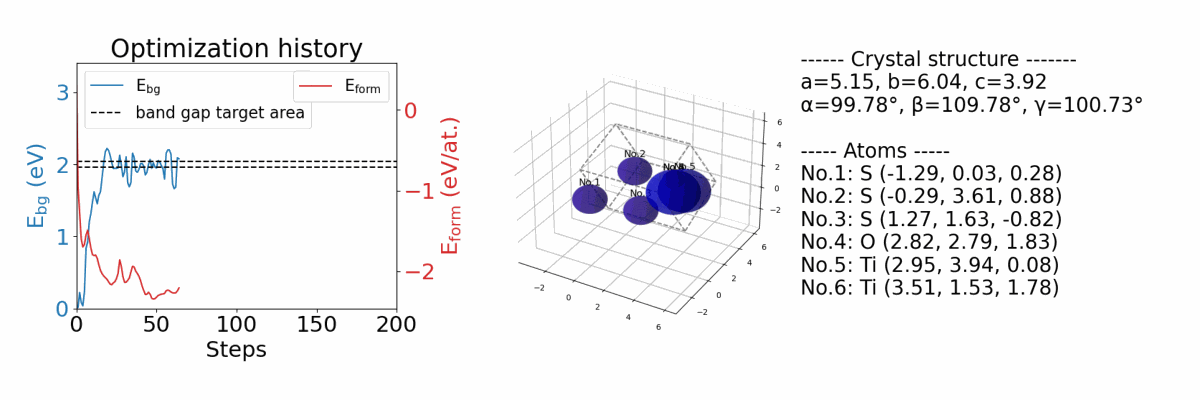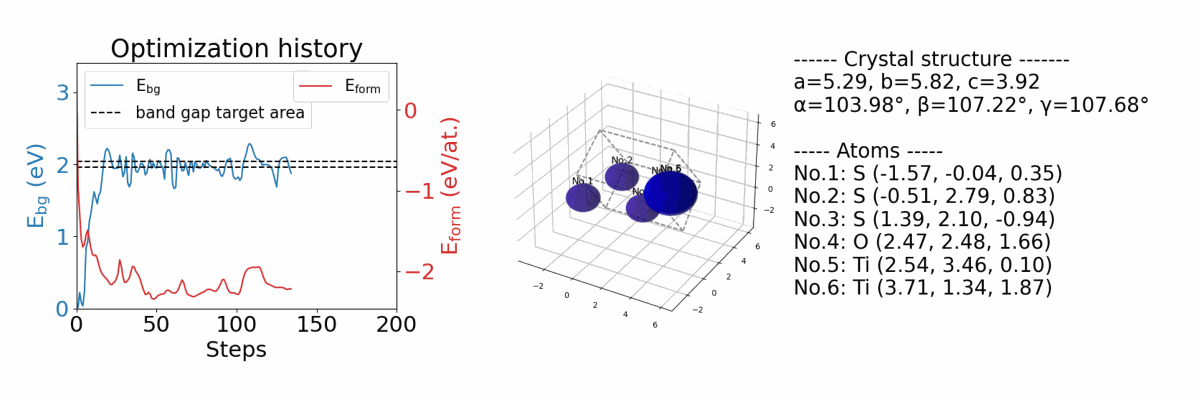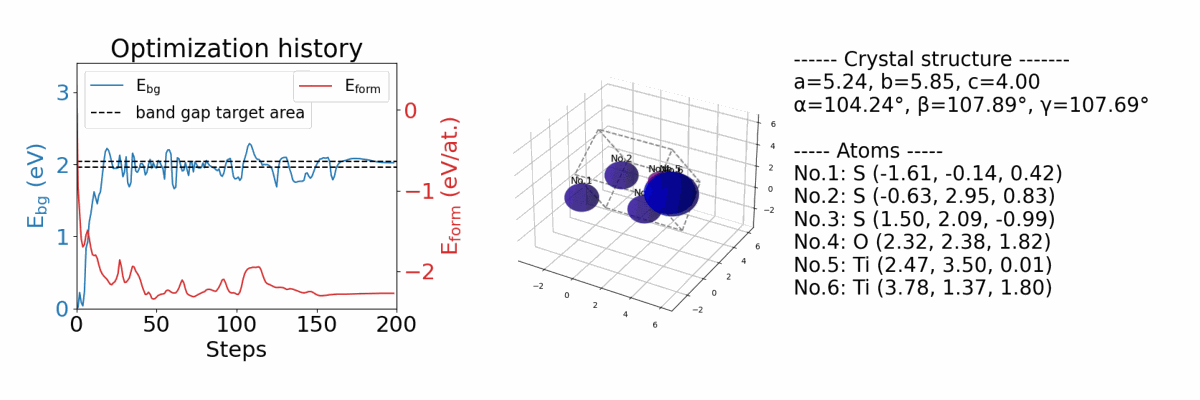
.results/{exp_name}/history_img.png |
an image file of optimization history. (if you optimize both band gap and formation energy) |
| column | description |
|---|---|
original_fnames |
file names of initial structures in the directory of dir_path
|
batch_abc |
values of lengths of crystal vectors |
batch_angle |
values of angles of crystal vector |
*_onehot |
optimized values with one-hot atomic distributions. e.g., bandgap_onehot
|
loss_*_onehot |
loss values after optimization with one-hot atomic distributions. e.g., loss_bandgap_onehot
|
*_success |
whether to meet its criterion |
valid_structure |
whether to meet both is_neutral and minbond_less_than_0.5
|
is_neutral |
electrically neutrality |
minbond_less_than_0.5 |
whether the all bonds are larger than 0.5Å |
is_neutral |
electrically neutrality |
ox_states |
oxidation numbers if the structure is electrically neutral. The order is the same as elements. |
perov_success |
whether to meet both tolerance_success and perov_coords
|
tolerance_success |
whether tolerance value |
perov_coords |
whether displacements of fractional coordinates are within |
success |
overall success. If the following conditions are met at the same time: *_success and valid_structure for non-perovskite optimization; *_success, valid_structure and perov_success for perovskite optimization |
A setting for S(Cry) on Table 2 in our paper.
main_experiment(
dir_path = './initial_crystal_candidates/structures_from_MEGNet', # directory of initial crystal structures
mask_data_path = './mask_arrays/mask_dict_super_common.npz', # atomic mask array
radii_data_path = None, # ionic radii array
exp_name = 'S_Cry_random', # experiment name.
model_name = 'Crystalformer' , # model name. 'Crystalformer' or 'ALIGNN'
num_steps=200, # optimization steps
num_candidate=8, # The number of initial crystal structures
num_batch_crystal=8, # Batch size to optimize
# prediction model, loss and criteria for each target values
prediction_loss_setting_dict ={
'bandgap': {
'prediction_model':None,
'loss_function':{
'func_name':'GapLoss',
'margin':0.04, # (eV)
'target_bandgap':2.0, # (eV)
'mode':'band',
'coef':1.0, # coefficient for the loss
},
'criteria_max':2.04, # optimized values should be less than this value
'criteria_min':1.96, # optimizedvalues should be more than this value
},
'e_form':{
'prediction_model':None,
'loss_function':{
'func_name':'FormationEnegryLoss',
'e_form_min':None,
'e_form_coef':1.0, # coefficient for the loss
},
'criteria_max':-0.5, # optimized values should be less than this value
'criteria_min':None, # optimizedvalues should be more than this value
}
},
atomic_dictribution_loss_setting_dict = {},
site_atom_oxidation_dict = {'Atom_settings': {}, 'use_ionic_radii':False},
limit_coords_displacement= None, # limiation for displacement of fractional coordindates
lattice_lr= 0.01, # learning rate for lattice (a,b,c,α,β,γ) optimization
coords_lr= 0.02, # learning rate for fractional coordinates
atom_lr= 0.6, # learning rate for atomic distribution and oxidation state configuration parameters
) The settings are almost the same as S(ALI) on the Table 3 in our paper, but ALIGNN_num_update_graphs is changed for a small demonstration. The file ./initial_crystal_candidates/random_perovskite_1 contains a randomly shaped site_atom_oxidation_dict{'Atom_settings'}.
# ALIGNN / perovskite structure
main_experiment(
dir_path = './initial_crystal_candidates/random_perovskite_1', # directory of initial crystal structures
mask_data_path = './mask_arrays/ionic_mask_dict_super_common.npz', # atomic mask array
radii_data_path = './mask_arrays/ionic_radii_dict_super_common.npz', # ionic radii array
exp_name = 'S_ALI_perov',
model_name = 'ALIGNN' , #'Crystalformer' or 'ALIGNN'
num_steps=200,
num_candidate=16,
num_batch_crystal=16,#
prediction_loss_setting_dict ={
'bandgap': {
'alignn_model_name':"mp_gappbe_alignnn",
'prediction_model':None,
'loss_function':{
'func_name':'GapLoss',
'margin':0.04, # (eV)
'target_bandgap':3.5, # (eV)
'mode':'band',
'coef':1.0,
},
'criteria_max':3.54, # optimized values should be less than this value
'criteria_min':3.46, # optimizedvalues should be more than this value
},
'e_form':{
'alignn_model_name':"mp_e_form_alignnn",
'prediction_model':None,
'loss_function':{
'func_name':'FormationEnegryLoss',
'e_form_min':None,
'e_form_coef':1.0,
},
'criteria_max':-0.5, # optimized values should be less than this value
'criteria_min':None, # optimizedvalues should be more than this value
}
},
atomic_dictribution_loss_setting_dict = {
'tolerance':{
'loss_function':{
'func_name':'tolerance',
'coef':1.0,
'tolerance_range':(0.8, 1.0),
},
'criteria_max':1.0, # optimized values should be less than this value
'criteria_min':0.8, # optimizedvalues should be more than this value
},
},
site_atom_oxidation_dict = {
# site-wise specification of oxidation numbers
'Atom_settings': {
'Ba': {'ox_patterns':[2,1], 'site_id':0},
'Ti': {'ox_patterns':[4,2], 'site_id':1},
'O': {'ox_patterns':[-2,-1], 'site_id':2},
},
'use_ionic_radii':True # use ionic radius values to calculate tolerance t.
},
ALIGNN_num_update_graphs = 5,# frequency of graph updating.
limit_coords_displacement= 0.15, # limiation for displacement of fractional coordindates
perovskite_evaluation = True, # ionic radii array
angle_optimization = False, # disable optimization of angle
atom_lr= 0.00008,
lattice_lr= 0.5,
coords_lr= 0.002,
) In addition to the previous optimization: limit the element at the Ti site to either titanium or manganese, and change the optimization range of the angles to between 85° and 95°.
main_experiment(
exp_name = 'S_Cry_perov_ELM_limited',
model_name = 'Crystalformer' , #'Crystalformer' or 'ALIGNN' or 'Both
dir_path = './initial_crystal_candidates/random_perovskite_2',
mask_data_path = './mask_arrays/ionic_mask_dict_super_common.npz',
radii_data_path = './mask_arrays/ionic_radii_dict_super_common.npz',
num_steps=num_steps,
num_candidate=len(os.listdir('./initial_crystal_candidates/random_perovskite_2')), # the number of initial structures
num_batch_crystal=len(os.listdir('./initial_crystal_candidates/random_perovskite_2')), # batch size
prediction_loss_setting_dict ={
'bandgap': {
'alignn_model_name':"mp_gappbe_alignnn",
'prediction_model':None,
'loss_function':{
'func_name':'GapLoss',
'margin':0.04, # (eV)
'target_bandgap':2.0, # (eV)
'mode':'band',
'coef':1.0,
},
'criteria_max': 2.04,
'criteria_min': 1.96,
},
'e_form':{
'alignn_model_name':"mp_e_form_alignnn",
'prediction_model':None,
'loss_function':{
'func_name':'FormationEnegryLoss',
'e_form_min':None,
'e_form_coef':1.0,
},
'criteria_max': -0.5, # optimized values should be less than this value
'criteria_min': None, # optimizedvalues should be more than this value
}
},
atomic_dictribution_loss_setting_dict = {
'tolerance':{
'loss_function':{
'func_name':'tolerance',
'coef':1.0,
'tolerance_range':(0.8, 1.0),
},
'criteria_max':1.0, # optimized values should be less than this value
'criteria_min':0.8, # optimizedvalues should be more than this value
},
},
site_atom_oxidation_dict = {
'Atom_settings': {
'Ba': {'ox_patterns':[2], 'site_id':0},
'Ti': {'element':['Mn', 'Ti'], 'ox_patterns':[4],'site_id':1}, # Limit the element at the Ti site to either titanium or manganese
'O': {'ox_patterns':[-2], 'site_id':2},
},
'use_ionic_radii':True
},
limit_coords_displacement= 0.15,
angle_range = (85, 95),
perovskite_evaluation = True,
atom_lr= 6.0,
lattice_lr= 0.01,
coords_lr= 0.02,
) We experimented to see if the crystal structure of metallic silicon with a zero band gap could be identified. Initially, we extracted structures from the MEGNet dataset that contained only one atom besides Si, using them as the initial structure. The atomic distribution was fixed with a one-hot vector indicating silicon, and only the lattice constants were optimized. The target properties for optimization were a zero band gap and formation energy minimization. We chose silicon structures from the MEGNet dataset with a band gap of 0 eV as the reference and compared these with the optimized structures that exhibited the lowest formation energy. See details for Section A.3 in our paper.
main_experiment(
exp_name = 'metaric_Silicon',
model_name = 'Crystalformer' ,
dir_path = './initial_crystal_candidates/one_atom_site_lattice_except_Si',
mask_data_path = './mask_arrays/ionic_mask_dict_super_common.npz',
radii_data_path = './mask_arrays/ionic_radii_dict_super_common.npz',
num_steps=num_steps,
num_candidate=len(os.listdir('./initial_crystal_candidates/one_atom_site_lattice_except_Si')),
num_batch_crystal=len(os.listdir('./initial_crystal_candidates/one_atom_site_lattice_except_Si')),
prediction_loss_setting_dict ={
'bandgap': {
'prediction_model':None,
'loss_function':{
'func_name':'GapLoss',
'margin':0.00, # (eV)
'target_bandgap':0.0, # (eV)
'mode':'band',
'coef':1.0,
},
'criteria_max':0.0, # optimized values should be less than this value
'criteria_min':0.0, # optimizedvalues should be more than this value
},
'e_form':{
'prediction_model':None,
'loss_function':{
'func_name':'FormationEnegryLoss',
'e_form_min':None,
'e_form_coef':1.0,
},
'criteria_max':-0.5, # optimized values should be less than this value
'criteria_min':None, # optimizedvalues should be more than this value
}
},
atomic_dictribution_loss_setting_dict = {},
site_atom_oxidation_dict = {
'Atom_settings': {
'*': {'element':'Si', 'site_id':-999} # '*' is wildcard. It means all elements are converted to Silicon.
},
'use_ionic_radii':False,
},
atom_lr = 0.0, # element is fixed to siliton
lattice_lr = 0.01,
coords_lr = 0.01,
neutral_check=False, # skip neutrality check
)
from jarvis.core.atoms import Atoms
import pandas as pd
import numpy as np
df = pd.read_csv('./results/metaric_Silicon/result.csv').sort_values(['loss_bandgap_onehot', 'e_form_onehot']).head(30)
d = np.load('./results/metaric_Silicon/result.npz', allow_pickle=True)
for lattice_id in df.index.values:
fname = f"Optimized_based_on_{d['original_fnames'][lattice_id]}"
atom = Atoms.from_poscar(os.path.join('./results/metaric_Silicon/poscar', fname))
print(f'initial lattice_id:{d["original_fnames"][lattice_id]} ', atom.lattice.abc, atom.lattice.angles, atom.coords," pred ef(eV):",df.loc[lattice_id,"e_form_onehot"])Formation energy optimization
main_experiment(
dir_path = './initial_crystal_candidates/structures_from_MEGNet',
mask_data_path = './mask_arrays/mask_dict_super_common.npz',
radii_data_path = None,
exp_name = 'S_Cry_random_ef',
model_name = 'Crystalformer' ,
num_steps=200,
num_candidate=8,
num_batch_crystal=8,
prediction_loss_setting_dict ={
'e_form':{
'alignn_model_name':"mp_e_form_alignnn",
'prediction_model':None,
'loss_function':{
'func_name':'FormationEnegryLoss',
'e_form_min':None,
'e_form_coef':1.0,
},
'criteria_max': -0.5, # optimized values should be less than this value
'criteria_min': None, # optimizedvalues should be more than this value
},
},
atomic_dictribution_loss_setting_dict = {},
site_atom_oxidation_dict = {'Atom_settings': {}, 'use_ionic_radii':False},
atom_lr= 6.0,
lattice_lr= 0.01,
coords_lr= 0.02,
) band gap optimization
# Crystalformer / random structure
main_experiment(
dir_path = './initial_crystal_candidates/structures_from_MEGNet',
mask_data_path = './mask_arrays/mask_dict_super_common.npz',
radii_data_path = None,
exp_name = 'S_Cry_random_bg',
model_name = 'Crystalformer' ,
num_steps=200,
num_candidate=8,
num_batch_crystal=8,
prediction_loss_setting_dict ={
'bandgap': {
'prediction_model':None,
'loss_function':{
'func_name':'GapLoss',
'margin':0.04, # (eV)
'target_bandgap':2.0, # (eV)
'mode':'band',
'coef':1.0,
},
'criteria_max' : 2.04,
'criteria_min' : 1.96,
}
},
atomic_dictribution_loss_setting_dict = {},
site_atom_oxidation_dict = {'Atom_settings': {}, 'use_ionic_radii':False},
atom_lr= 6.0,
lattice_lr= 0.01,
coords_lr= 0.02,
) This project is licensed under the MIT License. Parts of this project include code from the National Institute of Standards and Technology (NIST). The NIST code is provided under the NIST license terms. For full license details, please see the LICENSE file.