*equal contribution
This repository contains the implementation of a deep learning framework for the differential diagnosis of dementia etiologies using multi-modal data.
Using data from
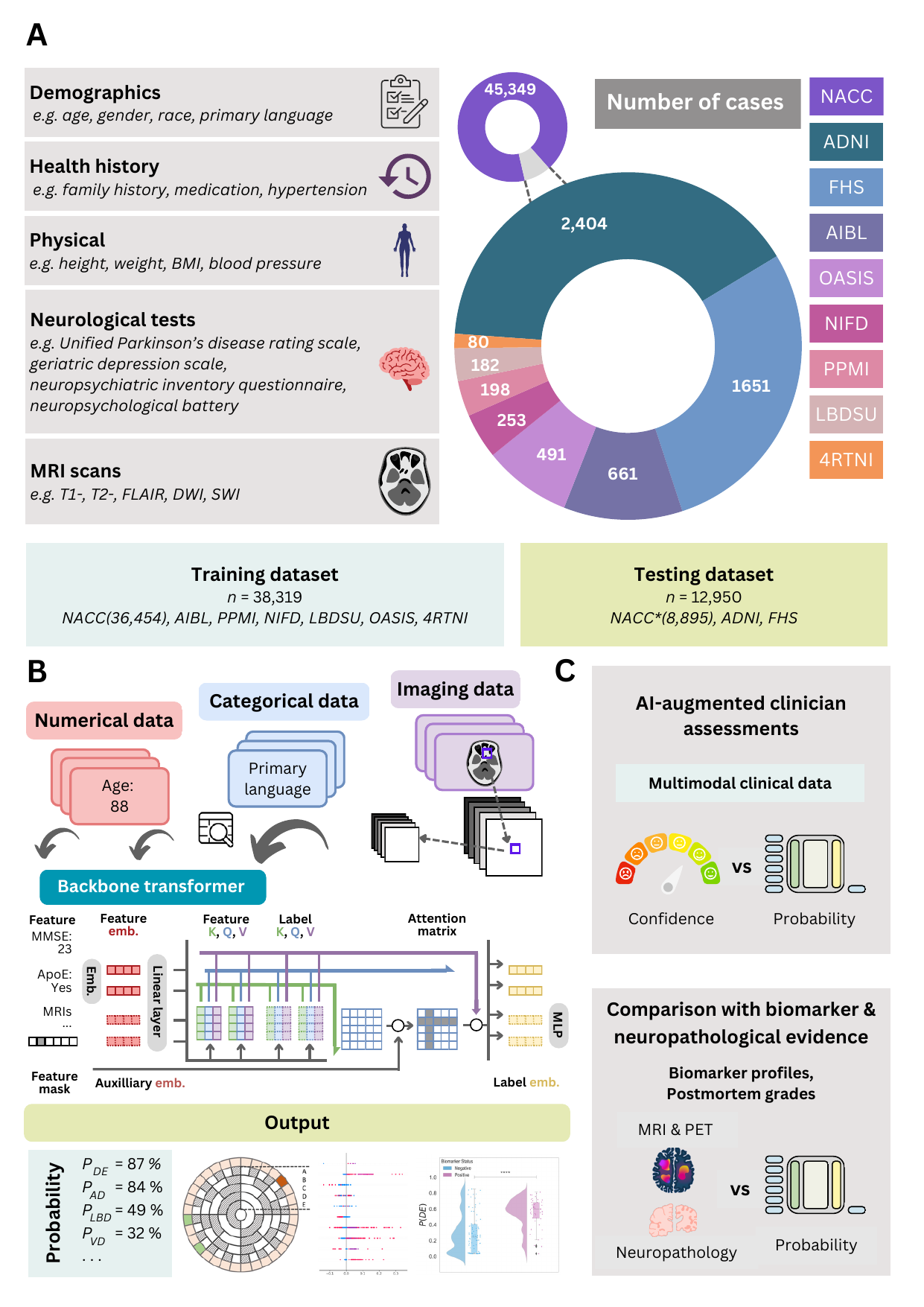
Figure 1: Data, model architecture, and modeling strategy. (a) Our model for differential dementia diagnosis was developed using diverse data modalities, including individual-level demographics, health history, neurological testing, physical/neurological exams, and multi-sequence MRI scans. These data sources whenever available were aggregated from nine independent cohorts: 4RTNI, ADNI, AIBL, FHS, LBDSU, NACC, NIFD, OASIS, and PPMI. For model training, we merged data from NACC, AIBL, PPMI, NIFD, LBDSU, OASIS and 4RTNI. We employed a subset of the NACC dataset for internal testing. For external validation, we utilized the ADNI and FHS cohorts. (b) A transformer served as the scaffold for the model. Each feature was processed into a fixed-length vector using a modality-specific embedding strategy and fed into the transformer as input. A linear layer was used to connect the transformer with the output prediction layer. (c) A distinct portion of the NACC dataset was randomly selected to enable a comparative analysis of the model’s performance against practicing neurologists. Furthermore, we conducted a direct comparison between the model and a team of practicing neuroradiologists using a random sample of cases with confirmed dementia from the NACC testing cohort. For both these evaluations, the model and clinicians had access to the same set of multimodal data. Finally, we assessed the model’s predictions by comparing them with pathology grades available from the NACC, ADNI, and FHS cohorts.
To setup the adrd package, run the following in the root of the repository:
pip install git+https://github.com/vkola-lab/nmed2024.gitThe tool was developed using the following dependencies:
- Python (3.11.7 or greater)
- PyTorch (2.1 or greater).
- TorchIO (0.15 or greater).
- MONAI (1.1 or greater).
- NumPy (1.24 or greater).
- tqdm (4.62 or greater).
- pandas (1.5.3 or greater).
- nibabel (5.0 or greater).
- matplotlib (3.7.2 or greater).
- shap (0.43 or greater).
- scikit-learn (1.2.2 or greater).
- scipy (1.10 or greater).
You can clone this repository using the following command:
git clone https://github.com/vkola-lab/nmed2024.gitThe training process consists of two stages:
All code related to training the imaging model with self-supervised learning is under ./dev/ssl_mri/.
Note: we used skull stripped MRIs to get our image embeddings. We have provided the script for skull stripping using the publicly available SynthStrip tool [2]. The code is provided under dev/skullstrip.sh.
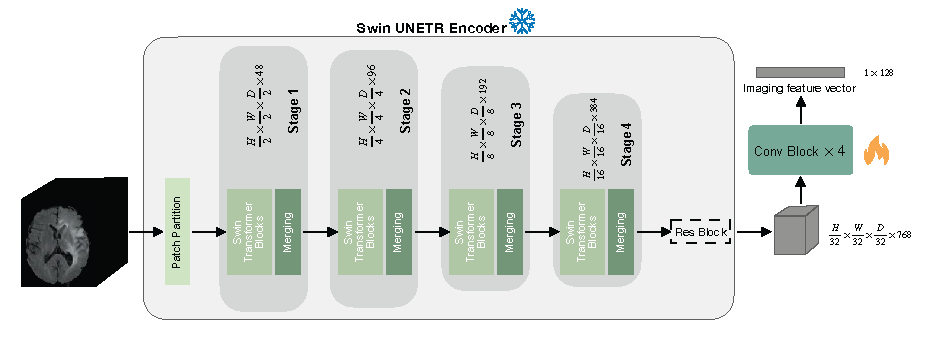
We trained started from the self-supervised pre-trained weights of the Swin UNETR encoder (CVPR paper [1]) which can be downloaded from this link. The checkpoint should be saved under ./dev/ssl_mri/pretrained_models/.
To finetune the pre-trained Swin UNETR on your own data, run the following commands:
cd dev/ssl_mri/
bash scripts/run_swinunetr.shThe code can run in a multi-GPU setting by setting --nproc_per_node to the appropriate number of available GPUs.
Once a finetuned checkpoint of the imaging model is saved, navigate to the repository's root directory and run dev/train.sh with the following changes in flag values:
img_net="SwinUNETR"
img_mode=2 # loads the imgnet, generates embeddings out of the MRIs input to the network, and saves them.
mri_type=ALL
emb_path="PATH/TO/SAVE/MRI/EMBEDDINGS"
Once image embeddings are saved, we train the backbone transformer on the multi-modal data.
Navigate to the repository's root directory and run dev/train.sh with the following changes in flag values:
img_net="SwinUNETREMB"
img_mode=1 # loads MRI embeddings and not the imgnet.
mri_type=SEQ
emb_path="PATH/TO/SAVED/EMBEDDINGS"All evaluation reports, including AUC-ROC curves, AUC-PR curves, confusion matrices, and detailed classification reports, were generated using the script dev/visualization_utils.py.
To make our deep learning framework for differential dementia diagnosis more accessible and user-friendly, we have hosted it on Huggingface Space. This interactive demo allows users to experience the power and efficiency of our model in real-time, providing an intuitive interface for uploading diagnostic information and receiving diagnostic predictions. Check out our Huggingface demo https://huggingface.co/spaces/vkola-lab/nmed2024 to see our model in action and explore its potential.
[1] Tang, Y., Yang, D., Li, W., Roth, H.R., Landman, B., Xu, D., Nath, V. and Hatamizadeh, A., 2022. Self-supervised pre-training of swin transformers for 3d medical image analysis. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (pp. 20730-20740).
[2] Hoopes, A., Mora, J.S., Dalca, A.V., Fischl, B. and Hoffmann, M., 2022. SynthStrip: Skull-stripping for any brain image. NeuroImage, 260, p.119474.