A project modelling the spread of infectious diseases via contact networks
Explore the docs »
View Demo
·
Report Bug
·
Request Feature
Table of contents
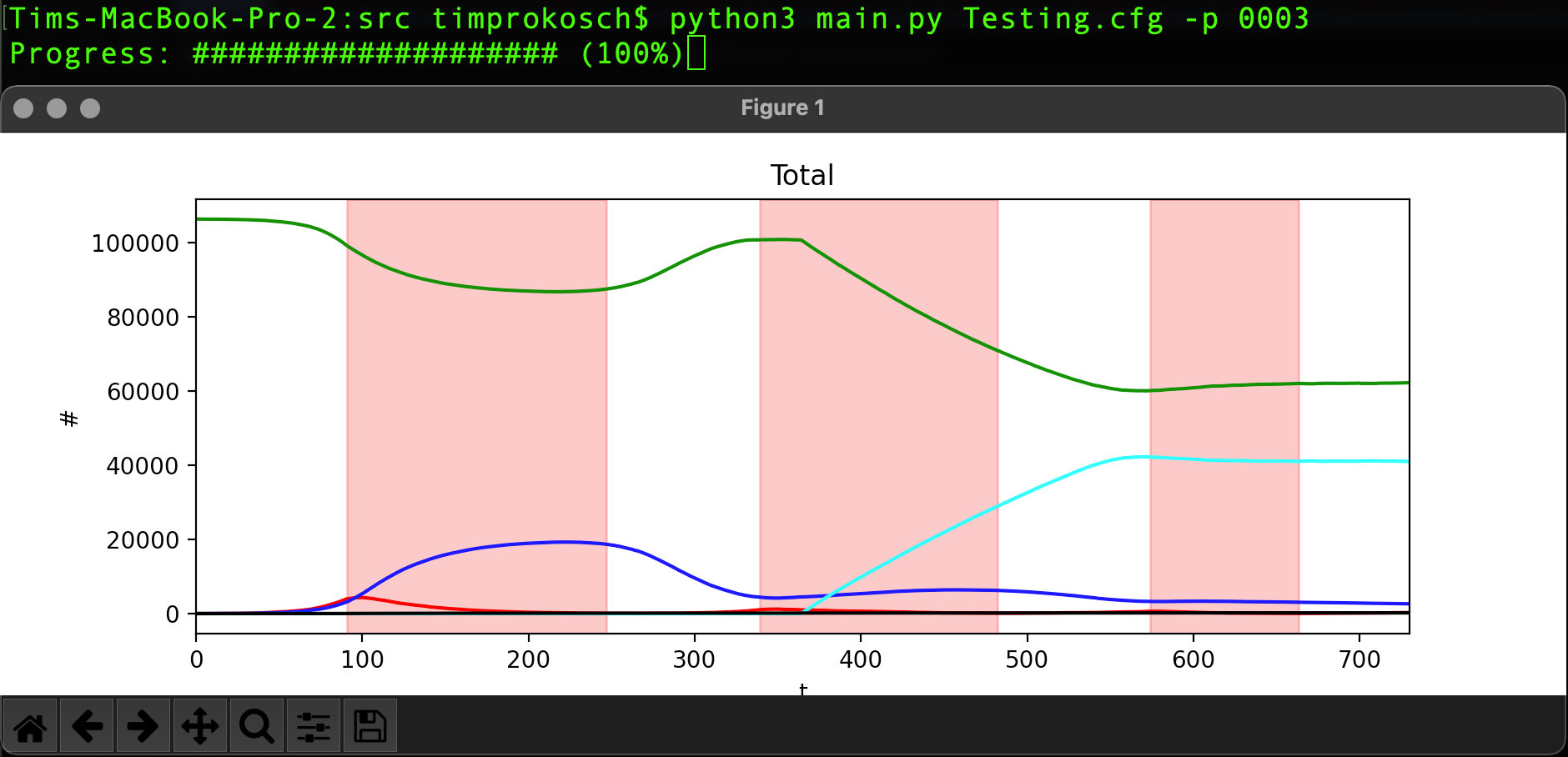
This project gives the user the ability to simulate customizable infection precesses using the many offered parameters that can be easily accessed (as seen here) and edited. Additionally, some post-processing methods are presented to analyze the outputted data, either directly after the simulation or at a later point in time.
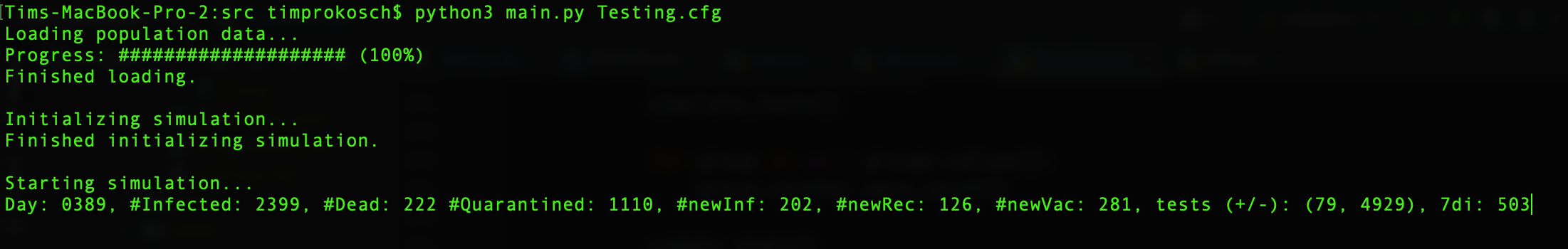
To start a simulation just use the python3 main.py SETTINGS_FILE_NAME.cfg command in your terminal from inside
the src folder of the project. To only execute the in the config file specified post-processing methods on the specified
population use python3 main.py SETTINGS_FILE_NAME.cfg -p SIMULATION_NUMBER.
For more examples, please refer to the Documentation
- More post-processing
- More heuristics
- More features
- Randomly picked interactions
- More measures like mask-duty
- Hospitalisation and infection severity
- Bigger populations (all of Germany?)
- Different virus variants
See the open issues for a full list of proposed features (and known issues).
Distributed under the MIT License. See LICENSE.txt for more information.
Tim Prokosch - prokosch@rhrk.uni-kl.de
Tobias Roth - tproth@rhrk.uni-kl.de
Project Link: https://github.com/AlbertEMC2Stein/COVID19-CNS