This script allows you to convert a samples.txt file generated
from the Emmanuel Caruyer's web application
to a tensorXXX.dat file you can use for DTI sequences on a GE MRI System.
As usual it's "AS IS" and for research only !
Please read the Tests part of this README carefully.
Please kindly cite the following relevant article when you use the sampling scheme :
Emmanuel Caruyer, Christophe Lenglet, Guillermo Sapiro, Rachid Deriche. Design of multishell sampling schemes with uniform coverage in diffusion MRI. Magnetic Resonance in Medicine, Wiley, 2013, 69 (6), pp. 1534-1540. http://dx.doi.org/10.1002/mrm.24736
This installation was tested on a SIGNA Premier system (RX29.1).
Download your sample scheme from the Emmanuel Caruyer's web application
Get the script and the file samples.txt to your home directory and open a terminal.
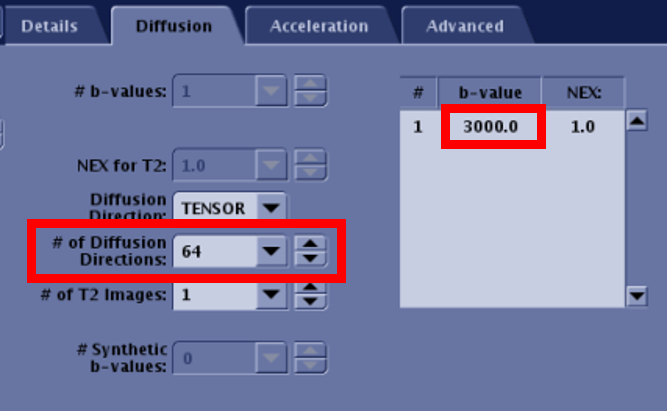
Generate the tensorXXX.dat from the samples.txt file (here with an example of 64 directions and 3 shells) :
python qspacesampling2ge.py samples.txt tensorXXX.dat 3000 2000 1000Copy and rename the tensorXXX.dat to /usr/g/bin/ (be careful to not erase previously existing tensor files) :
ls -l /usr/g/bin/tensor*
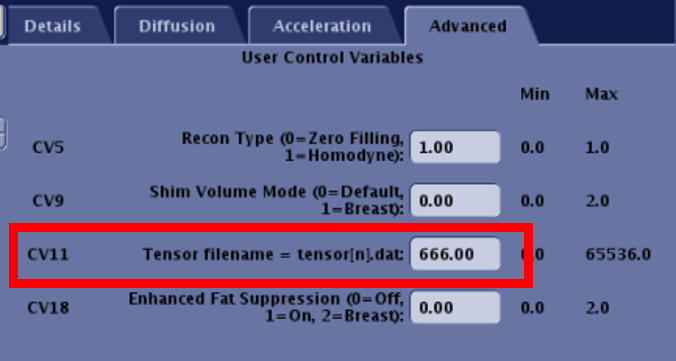
cp tensorXXX.dat /usr/g/bin/tensor666.datIn a DTI sequence :
- Set the number of direction to the number you setup in the Q-space-sampling scheme
- Set the b-value to the maximum b-value of your shell
- use the advanced panel to setup the tensor file number.
This script was tested on a 3T MRI SIGNA PREMIER system (MR29.1).
Using the same sampling described in the Usage section, here is the results of the comparison of the bvec/bval files obtained after converting the DICOM using dcm2niix and the original samples.txt file.
The results are given by the tests/test.py script (run it in the tests directory)
You can use this script with your own schemes like that :
python test.py --help
python test.py --samples samples.txt --bvec dcm2niix_nifti.bvec --bval dcm2niix_nifti.bval --bvalues 1000 2000 3000samples.txt is the file download from the Q-Space-Sampling web app. bvec/bval files are the ones created by dcm2niix when converting your dicom to nifti. The script need to know the expected b-values you setup. It will skip the bvec/bval for any b=0.
The u_x given by the sampling scheme and the x coordinate obtained in the bvec file are reversed in sign.
I choose to flip the sign of the x coordinate in the script to correct this.
There are some case where the b-values is slightly off. It's probably because of rounding values. Vectors coming from the q-space-sample app do not always have a norm of exactly 1. I choose to rescale the norm of the vector before generating the GE file and it seems that works.
Please check carefully the DICOM outputs if you use this script.