One network for each mood.
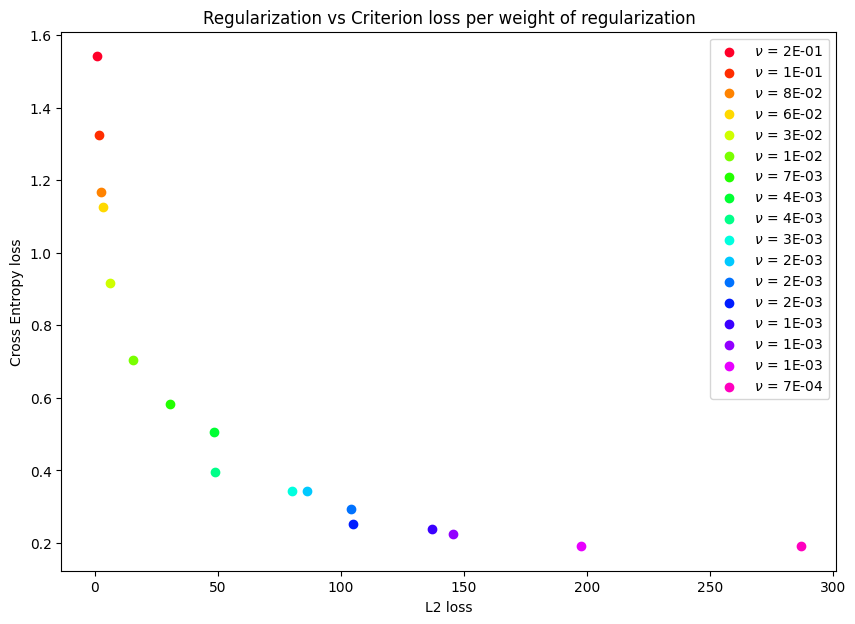
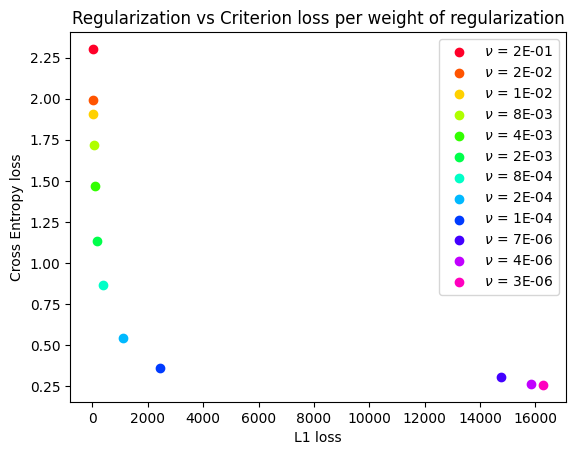
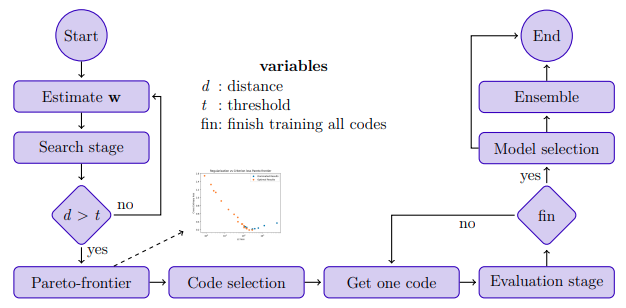
MOOD-NAS (Multi-objective Optimization Differentiable Neural Architecture Search) explore the common neglected conflicting objectives on NAS formulation. This way focuses on gradually adding regularization (complexity measure) strength to the model, thus filling an approximation of the Pareto frontier with efficient learning models exhibiting distinct trade-offs between error and model complexity. For a detailed description of technical details and experimental results, please refer to our Thesis: Multi-objective Differentiable Neural Architecture Search
Authors: Raphael Adamski, Marcos Medeiros Raimundo, Fernando Jose Von Zuben.
This code is based on the implementation of PC-DARTS which, in turn, is based on DARTS.
Smallest weights on above Pareto-frontier:
| Weight | Params(M) | Error(%) |
|---|---|---|
| ν = 3e-6 | 3.1 | 3.15 |
| ν = 4e-6 | 4.0 | 3.18 |
| ν = 7e-6 | 3.3 | 3.14 |
| ν = 1e-4 | 4.3 | 2.98 |
| ν = 2e-4 | 4.0 | 3.82 |
| Ensemble | 14.6 | 2.58 |
| AmoebaNet-B | 2.8 | 2.55 |
| DARTSV1 | 3.3 | 3.00 |
| DARTSV2 | 3.3 | 2.76 |
| SNAS | 2.8 | 2.85 |
| PC-DARTS | 3.6 | 2.57 |
To run the code, it is suggested a 12G memory GPU, but it can work with smaller sizes but slower (reduce the batch_size arg).
L2 loss
python multiobjective.py -o l2 cifar --set cifar10 --weight_decay 0.0
L1 loss
python multiobjective.py -o l1 cifar --set cifar10 --weight_decay 0.0
L1 loss with fixed value of l2
python multiobjective.py -o l1 cifar --set cifar10 --weight_decay 3e-4
Other parameters are compatible with python train_search.py original arguments.
Imagenet
python multiobjective.py -o l1 imagenet --weight_decay 3e-4
Cifar100
python multiobjective.py -o l1 cifar --set cifar100 --weight_decay 3e-4
Run the following tool to give you more information about the samples found during above search process like latency, FLOPs, params and useful plots.
export PYTHONPATH=$PYTHONPATH:..; \\
python tools/analyse_search_logs.py -l log.txt
The log.txt is inside output folder of multiobjective.py run.
The evaluation process simply follows PC-DARTS configuration. Moreover, we created a script to predict GPU memory consumption automatically to make training various codes (with different size and batch_size) easily. More information about this in the appendix.
python batch_train.py --archs CODE [CODE ...] --train_portion 1.0\\
--auxiliary \\
--cutout \\
--set {cifar10,cifar100} \\
The CODE should be a variable name on genotypes.py file (e.g. PC_DARTS_cifar or l2_loss_2e01). Other parameters are compatible with python train.py original arguments.
The --train_portion controls the train/validation portion. 0.9 for instance use 90% of data for training ans 10% for validation. For this case the validation is used only to log the accuracy in the validation set. No decision or stopping criterion is made based on it.
Run the following tool to give you more information about the models evaluated during above training process like latency, FLOPs, params and useful plots.
export PYTHONPATH=$PYTHONPATH:..; \\
python tools/analyse_train_logs.py -s search_log [search_log ...] -t train_log [train_log ...]
The search_log is inside output folder of multiobjective.py run.
The train_log is inside each output folder of batch_train.py run (each model evaluation create an evaluation log folder) or you can use the unified log file generated by batch_train.py.
To create an ensemble from trained model run the following script:
python ensemble.py --models_folder log_folder \\
--calculate \\
--auxiliary \\
--cutout \\
--set {cifar10,cifar100} \\
The log_folder should be a path to a folder containing evaluation log sub folders. --calculate with calculate the weight of the models given the training set metrics. Other parameters are compatible with python train.py original arguments.
- All scripts from PC-DARTS were update and are now compatible with
python3+andpytorch1.8.1+. We ran all expriments on aTesla V100GPU. - All Pareto optimal codes (Genotypes) found on the search stage (using all three cases) are available at
genotypes.pyfile.
Partial Channel Connections for Memory-Efficient Differentiable Architecture Search
Differentiable Architecture Search
If you use our code in your research, please cite our Thesis accordingly.
@mastersthesis{
Adamski2022,
author = {Adamski, Raphael and Raimundo, Marcos Medeiros and Von Zuben, Fernando José},
title = "Multi-objective differentiable neural architecture search",
school = "School of Electrical and Computer Engineering, University of Campinas",
year = "2022",
url = {https://hdl.handle.net/20.500.12733/4482}
}
In order to create a naive estimator to memory consumption and avoid the cumbersome work to tune the batch_size for each architecture under evaluation, we create a dataset and a simple predictor using Polynomial Ridge regression.
import pickle
# load mode, sklearn is necessary
batch_model = pickle.load(open("batch_predict.pkl", "rb"))
batch_size = 200 # polite guess
params_in_millions=3.63 # PC-DARTS size
# predict memory consumption given the number of params and batch_size
consumption = batch_model.predict([[params_in_millions, batch_size]])python batch_size_predict.py --data csv_file
where the csv_file should has the same format of the batch_data.csv with columns 'model size' 'batch size' and 'GPU mb'