Protein Secondary Structure Prediction Tool using non-ML approaches (Chou-Fasman and GOR Algorithms). This project was created as a part of the course Programming For Scientists taken by Professor Phillip Compeau at Carnegie Mellon University, Department of Computational Biology.
main.go- Main file for the program. Contains the main function in which we call the algorithm-executing functions.datatypes.go- Custom datatypes used throughout the project.CFparameters.txt- Parameters file for Chou-Fasman prediction algorithm.GorParams/*- Parameter files for GOR prediction algorithm.aa_index_map.txt- Amino acid index map for Chou-Fasman algorithm.aa_index_map_gor.txt- Amino acid index map for the GOR algorithm.SSPTandSSPT.exe- Executable files for the program for Mac and Windows.3d_visualization_resources- Contains the pdbs and template html required for the 3d visualization.outputs- Contains the output files (2D plots, 3D html, and downloaded fastas of the ensembls provided).results- Contains the output from testing the accuracies of the GOR and Chou-Fasman predictions.results/ExpectedValues/*- Contains validation files from SECNET 2018 dataset.results/Output/results.csv- Contains the csv file containing the accuracies of the algorithms.
miscellaneous- Contains the miscellaneous files that are not required for the program to run, but were used in the development process.Results.ipynb- Jupyter notebook containing plots of our results for accuracies.
- To run the program, the gget package must be installed. Open terminal (create a new conda environment if needed) and run:
pip install --upgrade gget
- Then, clone the repository by executing the following. This will create a new folder called SSPT in your current directory.
git clone https://github.com/AndrewLutsky/SSPT.git
- Also, the go graphics package must be installed for the 2D visualization to work. Go to the cloned repository's directory and run:
go get -u github.com/fogleman/gg
- To build the executable file for the program, run:
go build
- Finally run:
./SSPT #for MacOS
SSPT.exe #for Windows
-
The program with first prompt you to choose the algorithm which you want to use for the predictions. Type in
1if you want to use Chou-Fasman, or2if you want to use GOR. Press enter. -
The program will prompt you to choose from three modes of operation:
array,FASTA, orDNA. Type in the mode you want to run (case-sensitive) and press enter.arraymode will prompt you to enter a comma separated list of ensembl IDs. The program will then download the FASTA and PDB corresponding to each ensembl ID and run the algorithm on each protein sequence.FASTAmode will prompt you to enter the path to a FASTA file. The program will then run the algorithm on each protein sequence in the file.DNAmode will prompt you to enter the path to a DNA file. The program will then perform ORF analysis on the DNA sequence and run the algorithm on each protein sequence in the file.
An Example could be:
1
array
ENST00000546271.1
- The ouputs will appear in the
outputsdirectory. The 3d visualization is supported only inarraymode. The 2d visualization is supported in all modes.
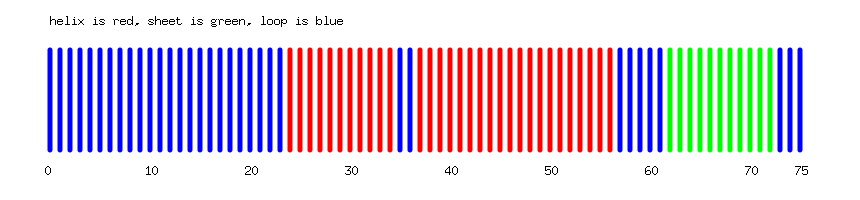
On Ubiquitin, the program outputs the following 2D visualization:
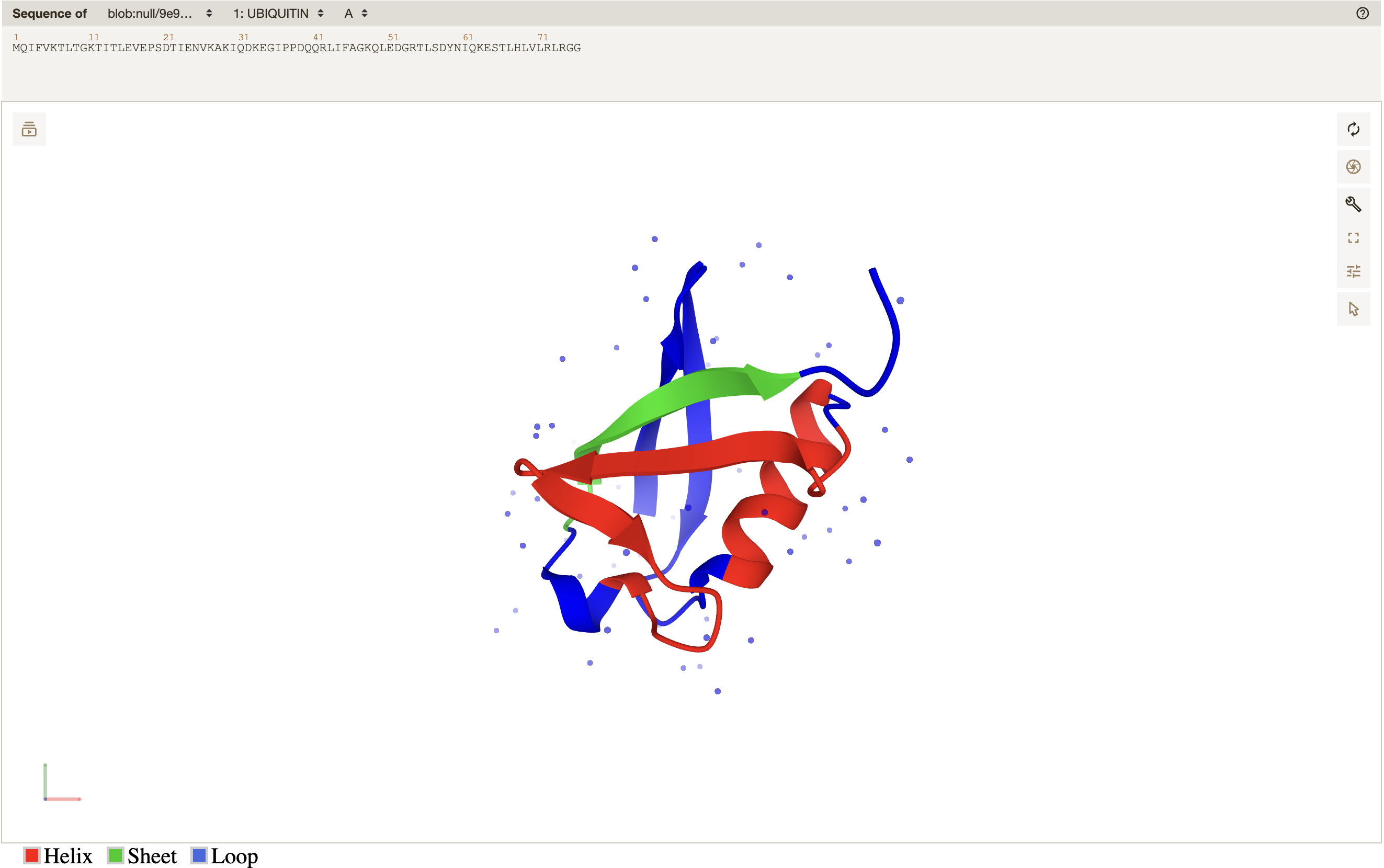
and the following 3D visualization:
- Jon Potter
- Shashank Katiyar
- Andrew Lutsky
- Rohit Nandakumar
- github.com/fogleman/gg
- github.com/pachterlab/gget
-
Jon Potter- Jon contributed to the project by constructing the code for most of the Chou-Fasman algorithm backend, everything along the pipeline from a string of amino acids to the collection of labeled features. Jon also laid the foundation for the GOR I method. Jon contributed to the presentation by constructing visual diagrams for the Chou Fasman algorithm slides. Jon contributed to the final report by writing about the Chou-Fasman algorithm and how it was applied to the problem at hand. Finally, Jon attended the weekly meetings to discuss the project’s direction and group work assignments. -
Andrew Lutsky- Andrew contributed to the overall project by implementing the identifying turns section of the CF algorithm as well as completing the overall implementation the GOR I algorithm and performing exploratory data analysis to visualize comparative accuracies of the two algorithms. He also attempted to implement GOR II using directional parameter restraints. Andrew contributed to the final report by writing about the GOR algorithm and how it was applied to the problem, DSSP classification, and the results section of the report.He also created visual representation of the walkthrough of the GOR algorithms. Additionally, Andrew attended the weekly meetings to discuss the project’s direction and group work assignments as well as performed last minute code commenting and overall cleanup. -
Rohit Nandakumar- Rohit contributed to the project by creating the input files for FASTA and CIF sequences. Additionally, Rohit assisted in the creation of the Chou-Fasman method, focusing on beta sheet identification. Rohit was heavily involved with creating testing functions for ten functions. In regards to the non-coding aspects of the project, Rohit wrote up the “Background” and “Future Advancements” sections of the paper and presentation. Lastly, Rohit attended weekly meetings with the group to discuss direction and progress. -
Shashank Katiyar- Shashank was heavily involved in creating the visualization tool for the project as well as implementing CLI workflow options. He tied together the various components of our project and provided a CLI for the user to interact with. Subsequently, he also created detailed instructions for the usage of the program in the Readme of the project and the demonstration video. Shashank also attended weekly meetings with the group to discuss direction and progress. Shashank contributed to the presentation by talking about the various different approaches taken to visualize the protein structure and to discuss future direction and progress.
https://drive.google.com/file/d/1Vhmn3ZukGElMmW9gib47cLjRKuTNiUfN/view?usp=sharing