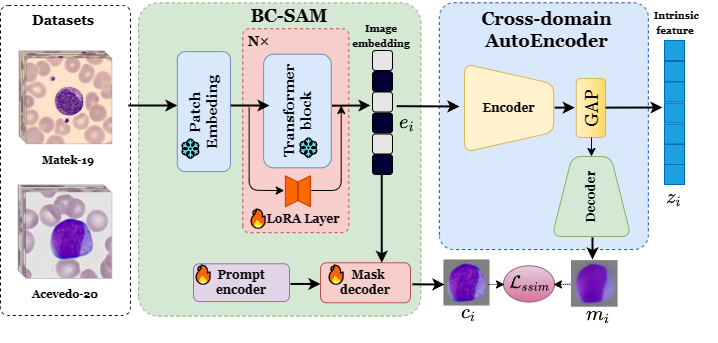
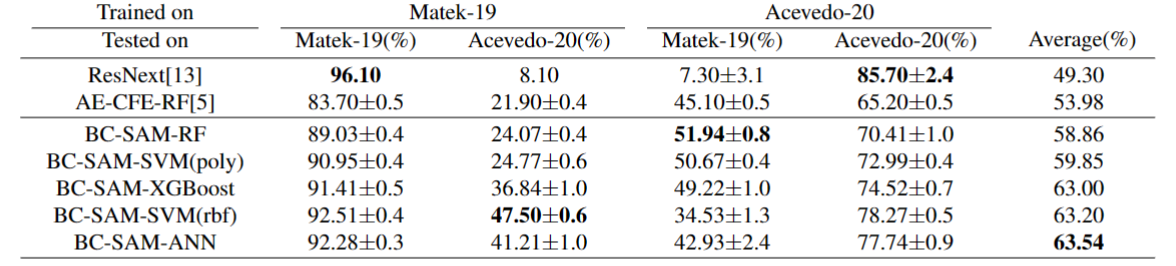
Official PyTorch implementation of Towards Cross-Domain Single Blood Cell Image Classification via Large-Scale LoRA-based Segment Anything Model (ISBI 2024).
We use two different white blood cells datasets to evaluate our method:
- The Matek-19 dataset consists of over 18,000 annotated white blood cells from 100 acute myeloid leukameia patients. It is published by Matek et al. Human-level recognition of blast cells in acute myeloid leukaemia with convolutional neural networks.
- The Acevedo-20 dataset has over 17,000 images which is published by Acevedo et al. A dataset of microscopic peripheral blood cell images for the development of automatic recognition systems.
cd BC-SAM
conda create -n BCSAM python=3.9
conda activate BCSAM
pip install torch==2.0.1+cu118 torchvision==0.15.2+cu118 --index-url https://download.pytorch.org/whl/cu118
pip install -r requirements.txt├── SAM
├── datasets
├── Bloodcell
├── Bloodcell_train.dat.gz
├── Bloodcell_test.dat.gzpython SAM/finetune_main.py \
--checkpoint model/sam_vit_b_01ec64.pth \
--datasets_dir SAM/datasets/Bloodcell \
--exit_path SAM/outputMatek-19
python SAM/sam_output.py
--path /path/to/dataset/Matek-19
--checkpoint_sam model/sam_vit_b_01ec64.pth
--checkpoint_path SAM/output/epoch_85.pth
--file_ext SAM/Matek-19.dat.gzAcevedo-20
python SAM/sam_output.py
--path /path/to/dataset/Acevedo-20
--checkpoint_sam model/sam_vit_b_01ec64.pth
--checkpoint_path SAM/output/epoch_85.pth
--file_ext SAM/Acevedo-20.dat.gzHere, --path is the path of the source dataset.
python train.py \
--datasets_dir SAM
--model_exit model/Here, --datasets_dir is the path of features and masks extracted from SAM.
Run feature/classification.ipynb to get the classification results.