In this study, a deep convolutional neural network was trained to classify single lead ECG waveforms as either Normal Sinus Rhythm, Atrial Fibrillation, or Other Rhythm. The study was run in two phases, the first to generate a classifier that performed at a level comparable to the top submission of the 2017 Physionet Challenge, and the second to extract class activation mappings to help better understand which areas of the waveform the model was focusing on when making a classification.
The convolutional neural network has 13 layers, including dilated convolutions, max pooling, ReLU activation, batch normalization, and dropout. Class activation maps were generated using a global average pooling layer before the softmax layer. The model generated the following average scores, across all rhythm classes, on the validation dataset: precision=0.84, recall=0.85, F1=0.84, and accuracy=0.88.
In the 2017 Physionet Challenge, competitors were asked to build a model to classify a single lead ECG waveform as either Normal Sinus Rhythm, Atrial Fibrillation, Other Rhythm, or Noisy. The dataset consisted of 12,186 ECG waveforms that were donated by AliveCor. Data were acquired by patients using one of three generations of AliveCor's single-channel ECG device. Waveforms were recorded for an average of 30 seconds with the shortest waveform being 9 seconds, and the longest waveform being 61 seconds. The figure below presents examples of each rhythm class and the AliveCor acquisition device.
Download the training dataset training2017.zip and place all .mat files in deep_ecg/data/waveforms/mat and the labels.csv file in deep_ecg/data/labels.
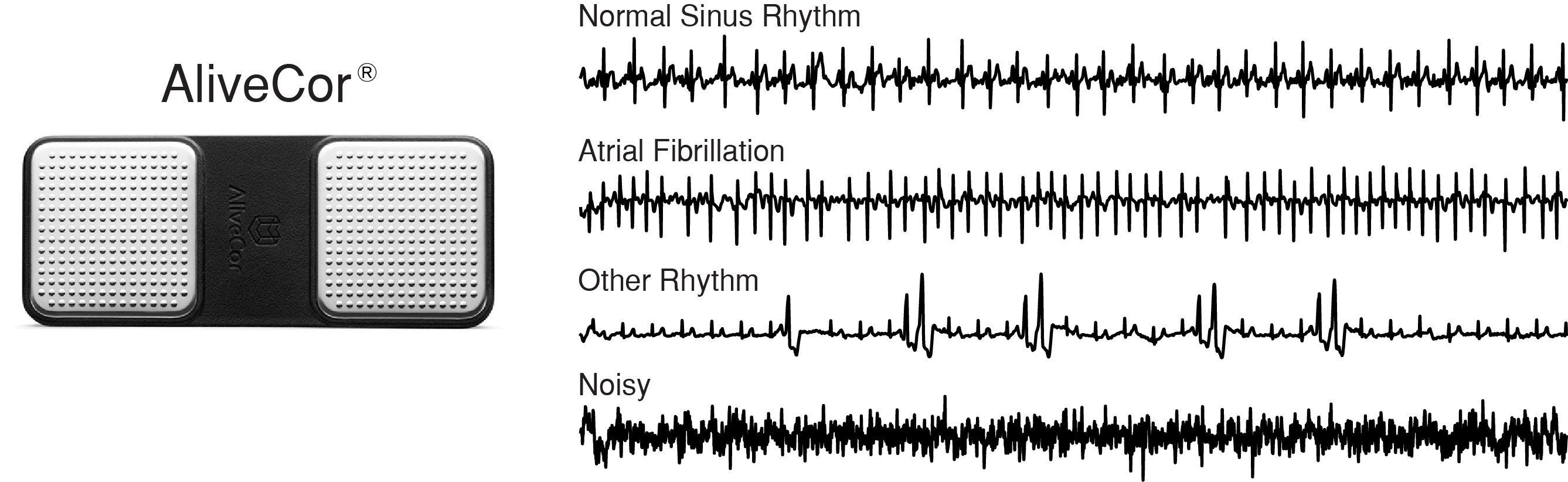 Left: AliveCor hand held ECG acquisition device. Right: Examples of ECG recording for each rhythm class,
Goodfellow et al. (2018).
Left: AliveCor hand held ECG acquisition device. Right: Examples of ECG recording for each rhythm class,
Goodfellow et al. (2018).
Zhou et al. (2016) demonstrate that convolutional neural networks trained for image classification appear to behave as object detectors despite information about the object's location not being part of the training labels (no bounding box annotations). Zhou et al. (2016)'s formulation was designed for analysis of images whereas our application is time series. For our application, the class activation map for a particular rhythm class was used to indicate the discriminative temporal regions, not spatial regions, used by the convolutional neural network to identify that rhythm class. Our work is a direct adaptation of Zhou et al. (2016)'s formulation for time series data.
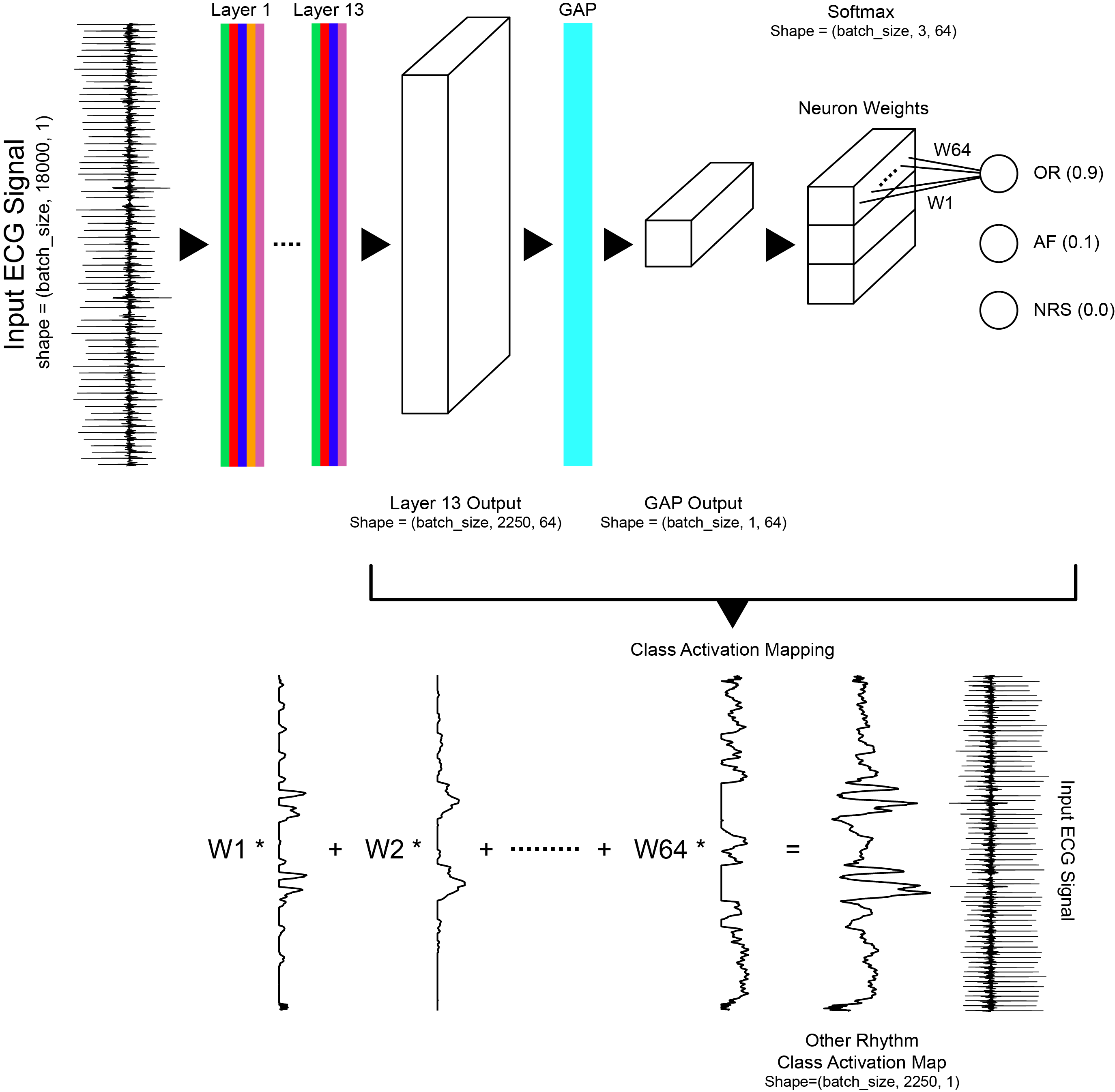 Class activation map formulation, Goodfellow et al. (2018).
Class activation map formulation, Goodfellow et al. (2018).
- Goodfellow, S. D., A. Goodwin, R. Greer, P. C. Laussen, M. Mazwi, and D. Eytan, Towards understanding ECG rhythm classification using convolutional neural networks and attention mappings, Machine Learning for Healthcare, Aug 17–18, 2018, Stanford, California, USA.
-
The Hospital for Sick Children
Department of Critical Care Medicine
Toronto, Ontario, Canada -
Laussen Labs
www.laussenlabs.ca
Toronto, Ontario, Canada
-
B. Zhou, A. Khosla, A. Lapedriza, A. Oliva, and A. Torralba. Learning Deep Features for Discriminative Localization. CVPR, 2016. DOI
-
Goodfellow, S. D., A. Goodwin, R. Greer, P. C. Laussen, M. Mazwi, and D. Eytan (2018), Atrial fibrillation classification using step-by-step machine learning, Biomed. Phys. Eng. Express, 4, 045005. DOI: 10.1088/2057-1976/aabef4
@conference{goodfellow_mlforhc_2018,
author = {S. D. Goodfellow and A. Goodwin and R. Greer and P. C. Laussen and M. Mazwi and D. Eytan},
title = {{Towards understanding ECG rhythm classification using convolutional neural networks and attention mappings}},
booktitle = {Proceedings of Machine Learning for Healthcare 2018 JMLR WC Track Volume 85, Aug 17–18, 2018, Stanford, California, USA},
year = 2018
}