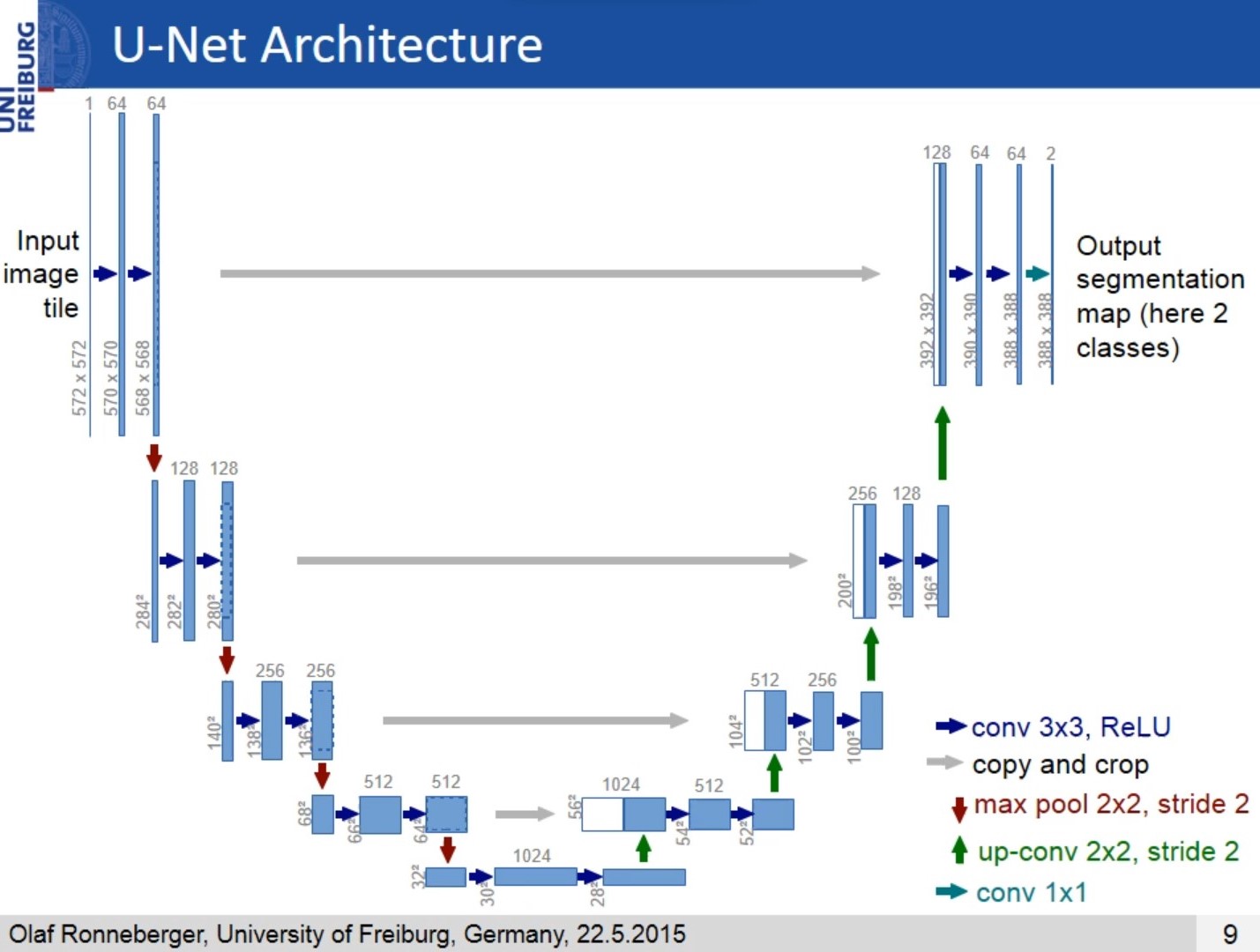
The architecture was inspired by U-Net: Convolutional Networks for Biomedical Image Segmentation.
The original dataset is from Kaggle: Br35H::Brain Tumor Detection 2020, and I've downloaded it and making PNG annotations from JSON annotation, and done the pre-processing.
Since the original dataset has 500 images for training and this number is small, I collected more data from different sources and generated more data using Flimimg.py. You can find it in /ImageSegmentation/MakeMoreData/ and use this dataset to train your custom model.
You can by using this site: Makesense annotate your data and use the JsonAnnotation_to_PNGAnnotation.py to make the PNG annotations. You can find it in /ImageSegmentation/AnnotationMaker/ .
This Dataset has been used to train these models.
This deep neural network is implemented with Keras functional API.
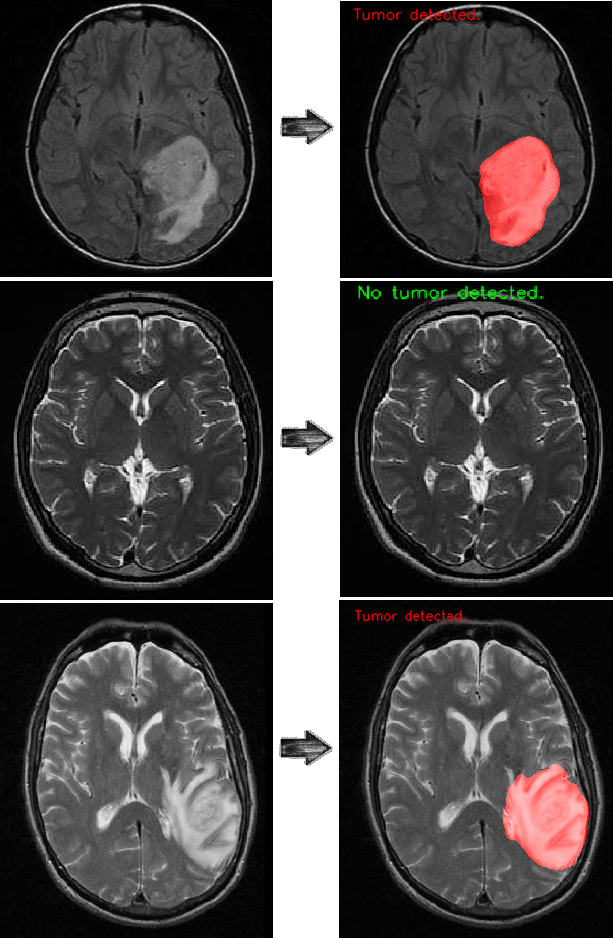
The U-net model is used to segment tumors in MRI images of the brain. After 10 epochs of training, the calculated accuracy is about 98%. Loss function for the training is basically just a binary crossentropy You can download my trained model from U-net
The CNN model is used to detect whether a tumor is there or not. After 15 epochs of training, the calculated accuracy is about 99.6%. Loss function for the training is basically just a binary crossentropy You can download my trained model from CNN
Use the trained model to do segmentation on test images. The result is satisfactory.