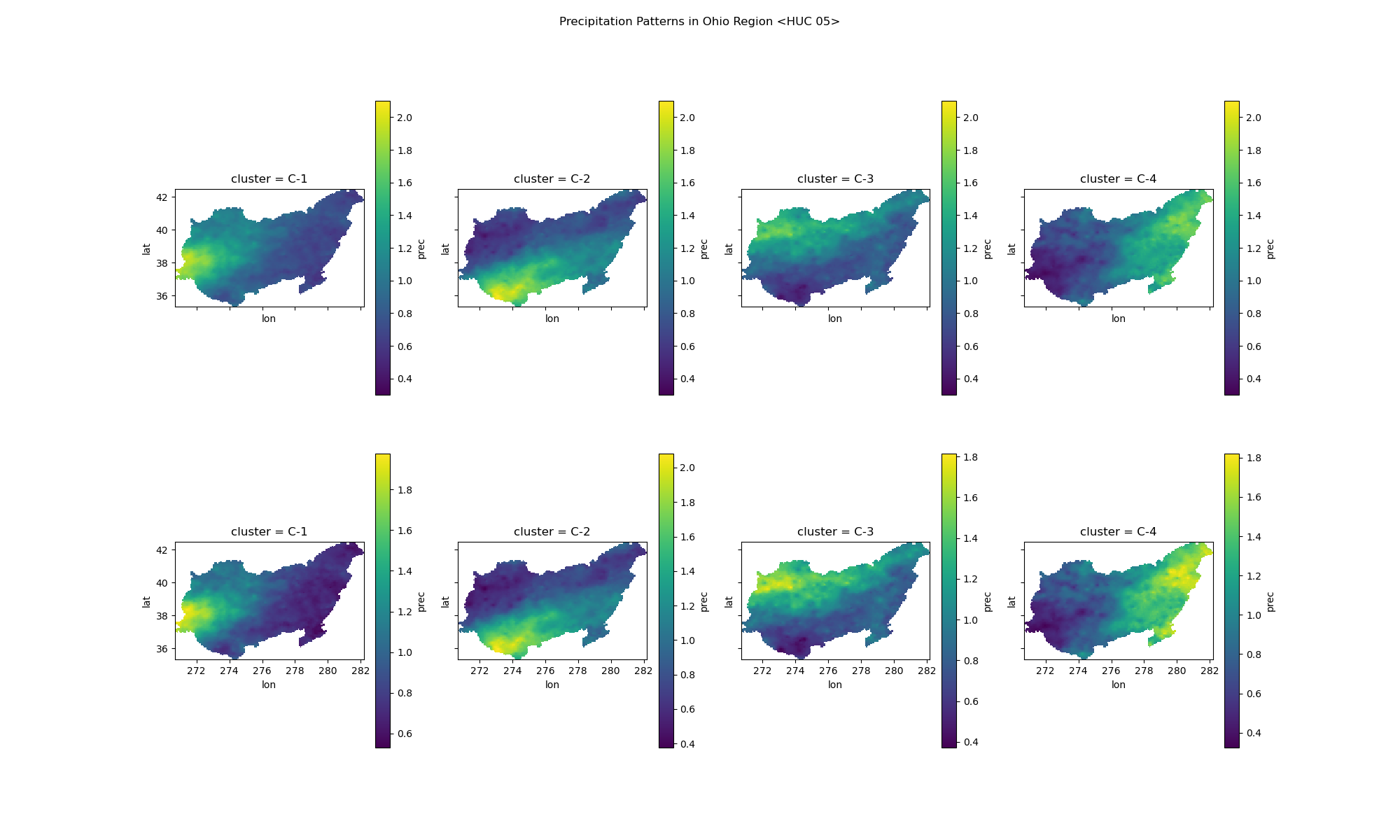
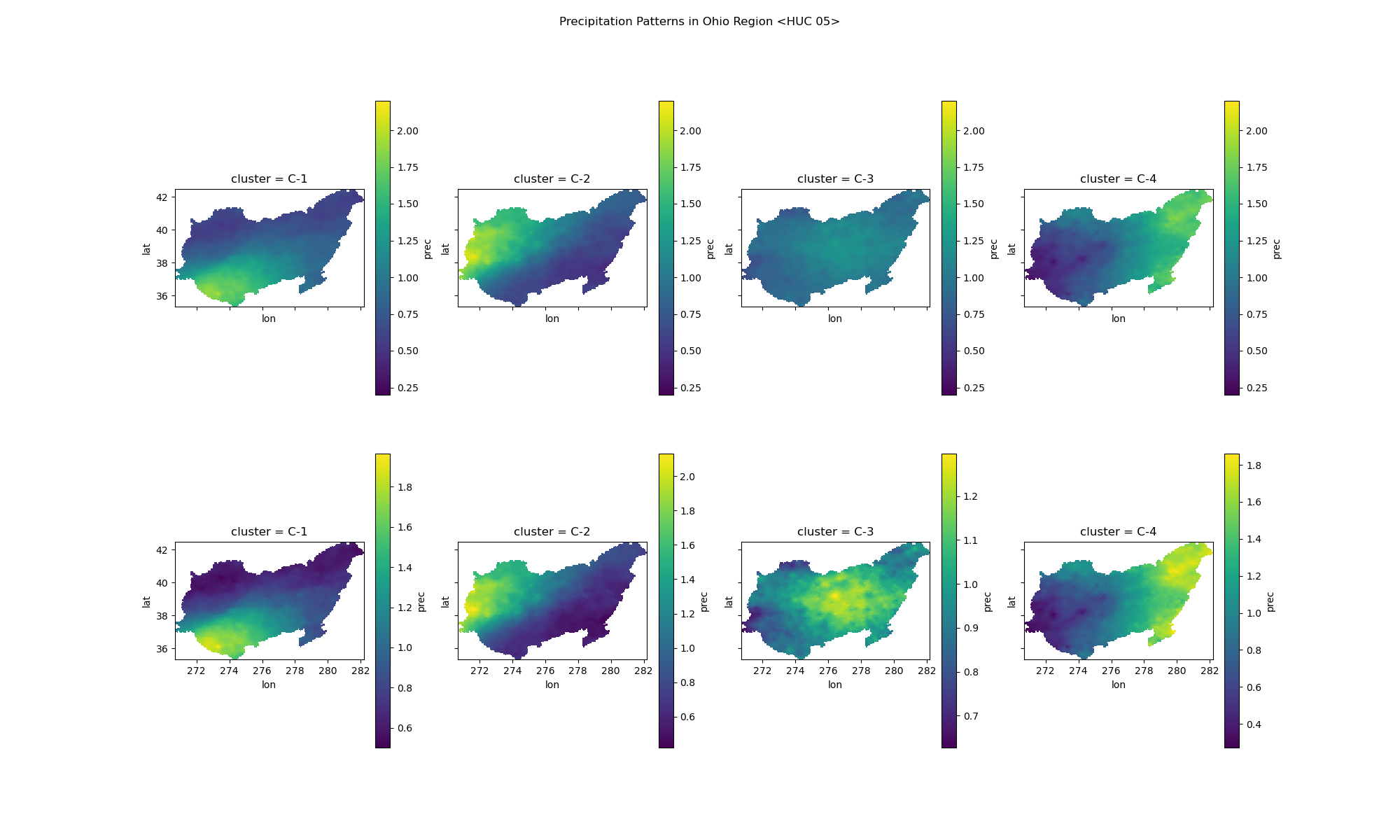
Identifies spatial patterns in extreme precipitation events.
Livneh datasets can be downloaded from: https://psl.noaa.gov/thredds/catalog/Datasets/livneh/metvars/catalog.html
Example link: https://psl.noaa.gov/thredds/fileServer/Datasets/livneh/metvars/prec.1915.nc Data is available from 1915 to 2011.
There is a download script (bash) that'll download the files for you in data directory, you need gnu parallel and curl for downloads, if you don't then it'll generate the links in the file data/livneh-files.txt, you can download it using your favourite download manager.
Download the watershed boundaries from USGS TNM Download (v2.0), direct link.
It's 4.8 GB to download and 7.1 GB after you unzip it.
The program has all the source codes inside the src directory, you can run it with python.
First install the requirements in requirements.txt
pip install -r requirements.txt
Then download the input files for Water Boundary Dataset and Livneh Dataset. See instructions above.
Run the CLI program with python
python -m src.locus -h
Will give you the usage instructions
usage: locus [-h] [-n NUM_DAYS] [-l] [-d] [-D] [-s {ams,pds,both}] [-c] [-e] [-a] [-p] [-b] HUCode
Analyse extreme precipitation pattern
positional arguments:
HUCode
options:
-h, --help show this help message and exit
-n NUM_DAYS, --num-days NUM_DAYS
Number of Days for ams/pds event
-l, --list-hucs List HUCodes in the given category and exit
-d, --details print basin details before processing
-D, --details-only print basin details and exit
-s {ams,pds,both}, --series {ams,pds,both}
ams or pds event to plot
-c, --calculate-weights
run the calculate_weights function
-e, --extract-annual-timeseries
run the extract_annual_timeseries function
-a, --ams-and-pds run the ams_and_pds function
-p, --plot-clusters run the plot_clusters function
-b, --batch-process run the batch_process function
https://github.com/Atreyagaurav/locus-code-usace
For example:
to run all the process for HUCode 02, for single day precipitation event:
python -m src.locus 02 -d -b -n 1
to list all HUC2 basins:
python -m src.locus 2 -l
For interactive Results visit: https://atreyagaurav.github.io/locus/index.html
For Ohio Region (HUC 05)
Clusters from AMS:
Clusters from PDS:
The program creates a directory with the HUCode as the name inside the data/output for data files (.csv, .nc, etc), and makes a directory with HUCode as name in the images for the plots (.png).
The files generated by each functions are discussed below.
Files: ids-and-weights.nc,ids.csv
Ids and Weights for each cells in the Livneh Dataset cropped to the given basin.
Files: prec.YYYY.csv (where YYYY is year from 1915-2011)
Daily timeseries of the basin averaged precipitation data for each year.
Files: {ams,pds}_Ndy_{series,grids}.csv where N is the num_days (duration of event considered)
AMS and PDS events for that basin for all the years. _series.csv has the basin averaged precipitation, while _grids.csv has the values of the precipitation in the grids inside the basin.
PDS is calculated with threhold value of minimum precipitation value of AMS events.
Files: {ams,pds}_Ndy.png where N is the num_days (duration of event considered)
Plot of the clusters made from the events data for the AMS list or PDS list. The images show clusters with individual scale on bottom, and global scale on top for comparision.
This is the runtime on Trinity (HUCode: 1203) on my laptop
- CPU: AMD Ryzen 7 6800U with Radeon Graphics (16) @ 2.700GHz,
- Storage: PCIe Gen 4 NVMe SSD Up to Seq Read: 4000MB/s, Write: 2000MB/s,
- RAM: 32 GB Dual Channel LPDDR5 up to 5500 MHz
On the first run, where it has to calculate everything:
*** CALCULATE_WEIGHTS
Time taken: 2.290 seconds ( 0.038 minutes)
*** EXTRACT_ANNUAL_TIMESERIES
Time taken: 86.598 seconds ( 1.4 minutes)
*** AMS_AND_PDS
Time taken: 23.125 seconds ( 0.39 minutes)
*** PLOT_CLUSTERS
Time taken: 3.401 seconds ( 0.057 minutes)
And then on the subsequent runs:
*** CALCULATE_WEIGHTS
Time taken: 0.002 seconds ( 2.9e-05 minutes)
*** EXTRACT_ANNUAL_TIMESERIES
Time taken: 0.075 seconds ( 0.0013 minutes)
*** AMS_AND_PDS
Time taken: 0.192 seconds ( 0.0032 minutes)
*** PLOT_CLUSTERS
Time taken: 3.503 seconds ( 0.058 minutes)
For North Branch Potomac (HUCode: 02070002) first runtime are as follows:
*** CALCULATE_WEIGHTS
Time taken: 0.104 seconds ( 0.0017 minutes)
*** EXTRACT_ANNUAL_TIMESERIES
Time taken: 83.067 seconds ( 1.4 minutes)
*** AMS_AND_PDS
Time taken: 9.011 seconds ( 0.15 minutes)
*** PLOT_CLUSTERS
Time taken: 2.308 seconds ( 0.038 minutes)
For Mid Atlantic Region <HUC 02>
*** CALCULATE_WEIGHTS
Time taken: 64.774 seconds ( 1.1 minutes)
*** EXTRACT_ANNUAL_TIMESERIES
Time taken: 91.243 seconds ( 1.5 minutes)
*** AMS_AND_PDS
Time taken: 22.159 seconds ( 0.37 minutes)
*** PLOT_CLUSTERS
Time taken: 22.557 seconds ( 0.38 minutes)
For Ohio Region <HUC 05>
*** CALCULATE_WEIGHTS
Time taken: 128.020 seconds ( 2.1 minutes)
*** EXTRACT_ANNUAL_TIMESERIES
Time taken: 91.441 seconds ( 1.5 minutes)
*** AMS_AND_PDS
Time taken: 18.734 seconds ( 0.31 minutes)
*** PLOT_CLUSTERS
Time taken: 32.160 seconds ( 0.54 minutes)
Seems like the extraction part is similar for all the basins, but the other ones vary by size. But it's reasonable time if you want to process a lot of basins.
The program is written to run everything without multiprocessing in the module itself, so multiple basins can be processed in parallel.
Use the --list-hucs command to get a list of codes for all basins, and then run the batch processing in parallel for them. You may want to filter the basins to only the ones that you want to process.
There are some basins that seem to be troublesome, you can add the --timeout flag on gnu parallel to kill processes that take too much time.
Here the first line makes the list of huc2 basins, edit it to the basins you want. Then run the second line that'll get the codes from that file and run locus on parallel, -j 4 will run 4 process in parallel at a given time, --timeout 200% will kill any process that takes more than double the median time to complete.
python -m src.locus -l 2 > basin-lists/huc2.txt
awk -F ' :' '{print $1}' < basin-lists/huc4.txt | parallel --timeout 200% -j 4 --bar 'python -m src.locus -d -b {0} >> reports/{0}.org'
You can also log the progress and resume from the where you left off. For more usage refer to gnu parallel manual.
The plots are a little weird, and I'm not that familiar with matplotlib, it seems to have a lot of extra border specially since we don't know how many clusters will be there, so I use imagemagick to trim the borders.
Here this example command will remove the excess padding and then add 20x20 padding on all sides
mogrify -trim +repage -bordercolor white -border 20x20 images/02/*
The output of the command can be piped, only the reporting texts from the main function of the src/locus.py is output to the stdout. The piped contents are in emacs-org format, which can be exported to any other formats (html,latex,pdf,etc) with emacs, if you do not have emacs you can use pandoc to convert it. Or you can modify the code to print markdown syntax instead, the output is simple so you only have to change all "*" in headings to "#".
If you need to print any debugging information that's not supposed to goto the report, print it in the stderr.
The locus server files are in server directory. The server data directory structure are as follows:
download/
thumbnails/
map.html
netcdfs-index.html
netcdfs-index.json
The generated netCDF files are in download directory, one file per grid value (cluster), and the netcdf files are converted into PNG images and saved in thumbnails. Following command can be used to convert the files in parallel using gnu parallel and gdal:
ls -1 download/*.nc | parallel --silent --bar echo 'gdaldem color-relief -alpha netcdf:{1} ../precip-color.txt thumbnails/{1.}.png'
As for the files:
- map.html: Interactive map showing the HUC boundaries and the clusters
- netcdfs-index.html: interactive page with tabulated data for the clusters
- netcdfs-index.json: tablulated data for the clusters in computer understandable languages for scripts
The Livneh Data Range is approximately from lat 25 to 53, and lon -125 to -67, which means it doesn't cover the following regions:
- Alaska Region <HUC 19> (-179.230, 51.157, 179.857, 71.440)
- Hawaii Region <HUC 20> (-178.431, 18.860, -154.753, 28.507)
- Caribbean Region <HUC 21> (-68.004, 17.624, -64.513, 18.567)
- South Pacific Region <HUC 22> (-176.674, -14.610, 166.711, 20.604)
The time reported can include other steps than the one you asked it to do, if that is needed. For example, if you asked for extracting the timeseries, then it'll calculate the weights if there is no weights, or if you ask for ams/pds it'll extract the timeseries.
HUC 09 takes a really long time (2.1 minutes in my laptop), I don't know why. Could be due to geometry? Need to check it out. Same with Great Lakes Region.