LEAP: Evolutionary Algorithms in Python
Written by Dr. Jeffrey K. Bassett, Dr. Mark Coletti, and Dr. Eric Scott
LEAP is a general purpose Evolutionary Computation package that combines readable and easy-to-use syntax for search and optimization algorithms with powerful distribution and visualization features.
LEAP's signature is its operator pipeline, which uses a simple list of functional operators to concisely express a metaheuristic algorithm's configuration as high-level code. Adding metrics, visualization, or special features (like distribution, coevolution, or island migrations) is often as simple as adding operators into the pipeline.
Get the stable version of LEAP from the Python package index with
pip install leap_ecThe easiest way to use an evolutionary algorithm in LEAP is to use the
leap_ec.simple package, which contains simple interfaces for pre-built
algorithms:
from leap_ec.simple import ea_solve
def f(x):
"""A real-valued function to optimized."""
return sum(x)**2
ea_solve(f, bounds=[(-5.12, 5.12) for _ in range(5)], maximize=True)The next-easiest way to use LEAP is to configure a custom algorithm via one
of the metaheuristic functions in the leap_ec.algorithms package. These
interfaces offer you a flexible way to customize the various operators,
representations, and other components that go into a modern evolutionary
algorithm.
Metaheuristics are usually defined by three main objects: a Problem, a
Representation, and a pipeline (list) of Operators.
Here's an example that applies a genetic algorithm variant to solve the
MaxOnes optimization problem. It uses bitflip mutation, uniform crossover,
and binary tournament_selection selection:
Python code for simple GA
from leap_ec.algorithm import generational_ea
from leap_ec import ops, decoder, probe, representation
from leap_ec.binary_rep import initializers
from leap_ec.binary_rep import problems
from leap_ec.binary_rep.ops import mutate_bitflip
pop_size = 5
final_pop = generational_ea(max_generations=10, pop_size=pop_size,
# Solve a MaxOnes Boolean optimization problem
problem=problems.MaxOnes(),
representation=representation.Representation(
# Genotype and phenotype are the same for this task
decoder=decoder.IdentityDecoder(),
# Initial genomes are random binary sequences
initialize=initializers.create_binary_sequence(length=10)
),
# The operator pipeline
pipeline=[
# Select parents via tournament_selection selection
ops.tournament_selection,
ops.clone, # Copy them (just to be safe)
# Basic mutation with a 1/L mutation rate
mutate_bitflip(expected_num_mutations=1),
# Crossover with a 40% chance of swapping each gene
ops.uniform_crossover(p_swap=0.4),
ops.evaluate, # Evaluate fitness
# Collect offspring into a new population
ops.pool(size=pop_size),
probe.BestSoFarProbe() # Print the BSF
])However, it may sometimes be necessary to have access to low-level details of an EA implementation, in which case the programmer can arbitrarily connect individual components of the EA workflow for maximum tailorability. For example:
Low-level example python code
from toolz import pipe
from leap_ec.individual import Individual
from leap_ec.decoder import IdentityDecoder
from leap_ec.context import context
import leap_ec.ops as ops
from leap_ec.binary_rep.problems import MaxOnes
from leap_ec.binary_rep.initializers import create_binary_sequence
from leap_ec.binary_rep.ops import mutate_bitflip
from leap_ec import util
# create initial rand population of 5 individuals
parents = Individual.create_population(5,
initialize=create_binary_sequence(4),
decoder=IdentityDecoder(),
problem=MaxOnes())
# Evaluate initial population
parents = Individual.evaluate_population(parents)
# print initial, random population
util.print_population(parents, generation=0)
# generation_counter is an optional convenience for generation tracking
generation_counter = util.inc_generation(context=context)
while generation_counter.generation() < 6:
offspring = pipe(parents,
ops.tournament_selection,
ops.clone,
mutate_bitflip(expected_num_mutations=1),
ops.uniform_crossover(p_swap=0.2),
ops.evaluate,
ops.pool(size=len(parents))) # accumulate offspring
parents = offspring
generation_counter() # increment to the next generation
util.print_population(parents, context['leap']['generation'])A number of LEAP demo applications are found in the the example/ directory of the github repository:
git clone https://github.com/AureumChaos/LEAP.git
python LEAP/examples/advanced/island_models.py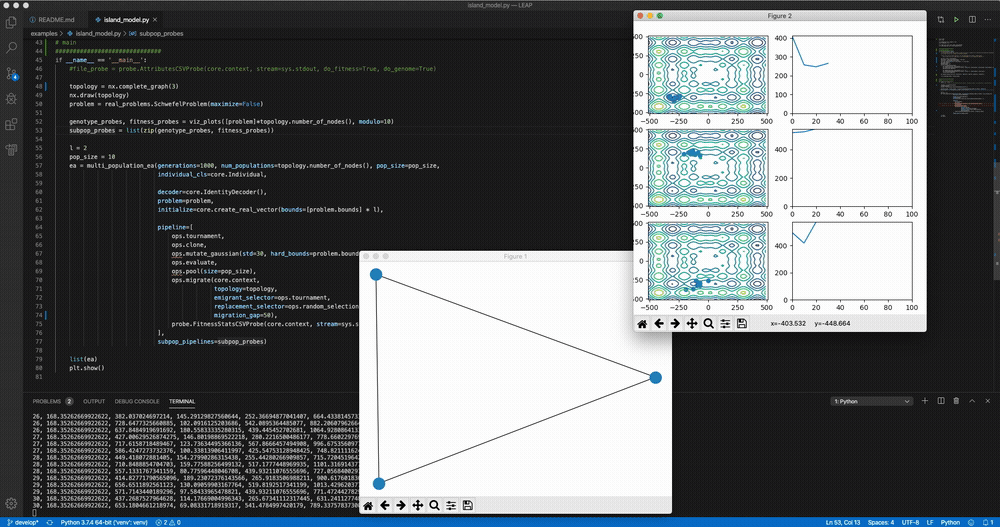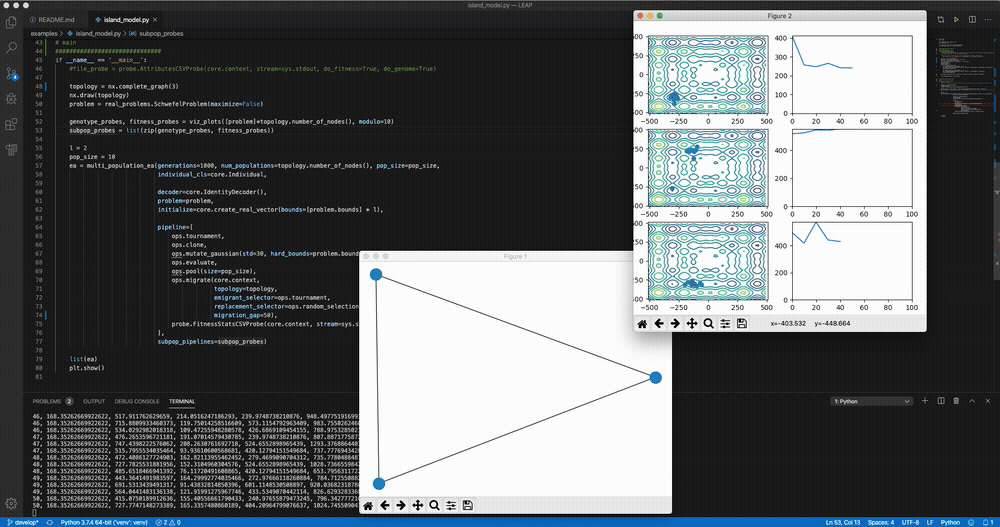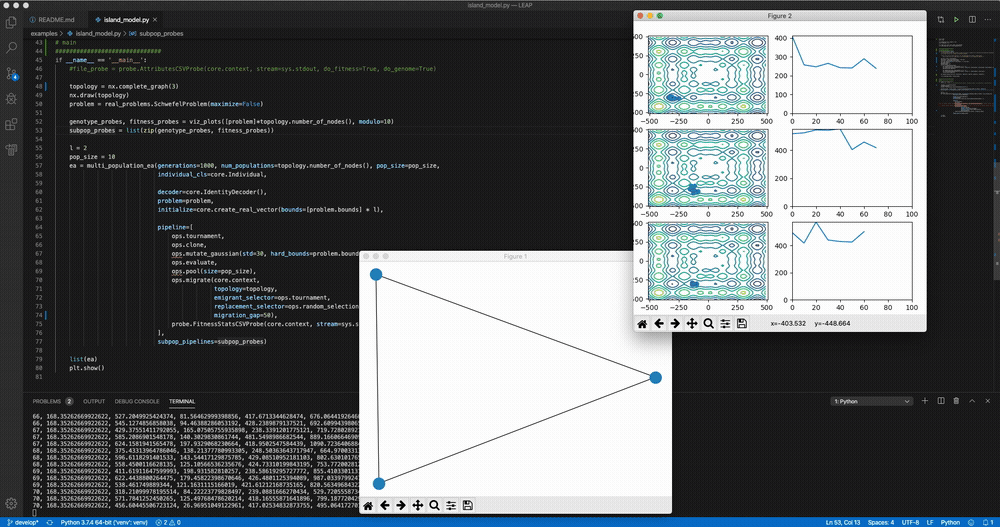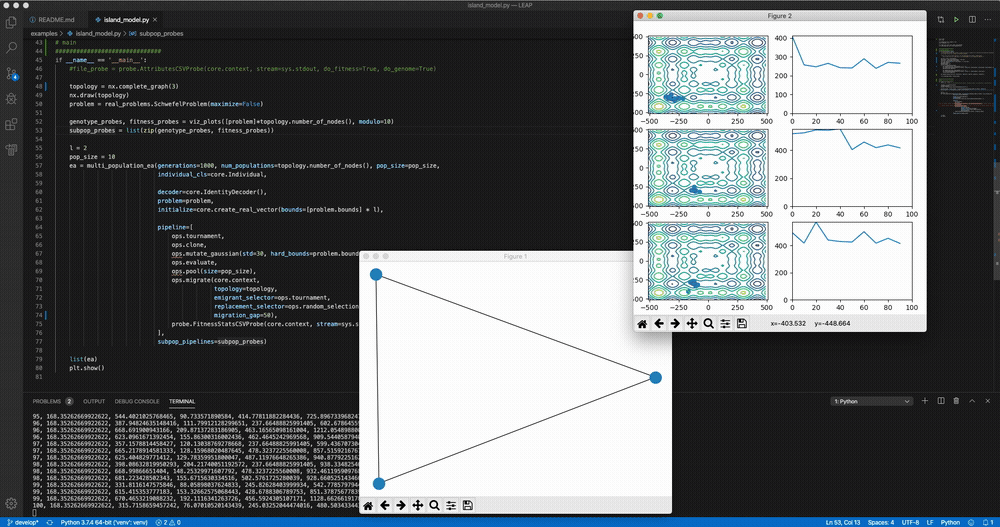 Demo of LEAP running a 3-population island model on a real-valued optimization problem.
Demo of LEAP running a 3-population island model on a real-valued optimization problem.
The stable version of LEAP's full documentation is over at ReadTheDocs.
If you want to build a fresh set of docs for yourself, you can do so after running make setup:
make doc
This will create HTML documentation in the docs/build/html/ directory. It might take a while the first time,
since building the docs involves generating some plots and executing some example algorithms.
To install a source distribution of LEAP, clone the repo:
git clone https://github.com/AureumChaos/LEAP.git
And use the Makefile to install the package:
make setupLEAP ships with a two-part pytest harness, divided into fast and slow tests. You can run them with
make test-fastand
make test-slowrespectively.