Coronary Artery Disease (CAD) is a major cause of death for men, women and people of all racial groups. In 2018, Egypt had 163,171 deaths from CAD, about 29% of the total deaths that year. According to WHO Egypt is ranked 15 on the world's rate of death from CAD with 271.9 deaths per 100,000 of population [1]. These numbers indicate the seriousness of the disease that Egypt is facing. Detecting this disease is time dependent and human error prone, Experienced Radiologists examine a patient's CT and calculate the volume of Calcifications found inside the patient’s heart. In this Paper, we propose a framework of Automated Deep learning Algorithms for the Quantification of the Calcification Volume from Low- dose Chest CT. Using two consecutive networks, the first is a Segmentation Network its main purpose is to identify our Region of Interest (ROI) which is the heart, each patient’s Specific ROI is then passed to the second network which is the Calcification Quantifier. Both networks Return a Segmented output. The Final output is then processed to roughly quantify the patient’s Calcification Volume which will help in his\her Therapy. The Results achieved by the Networks were assessed using the Dice Coefficient metric. Heart Segmentation output reached 90% Score. To reach out to the medical and scientific communities we then added this framework to a well- known Open-Source Application 3D Slicer, which will elaborate its usage and promote the research done in this area.
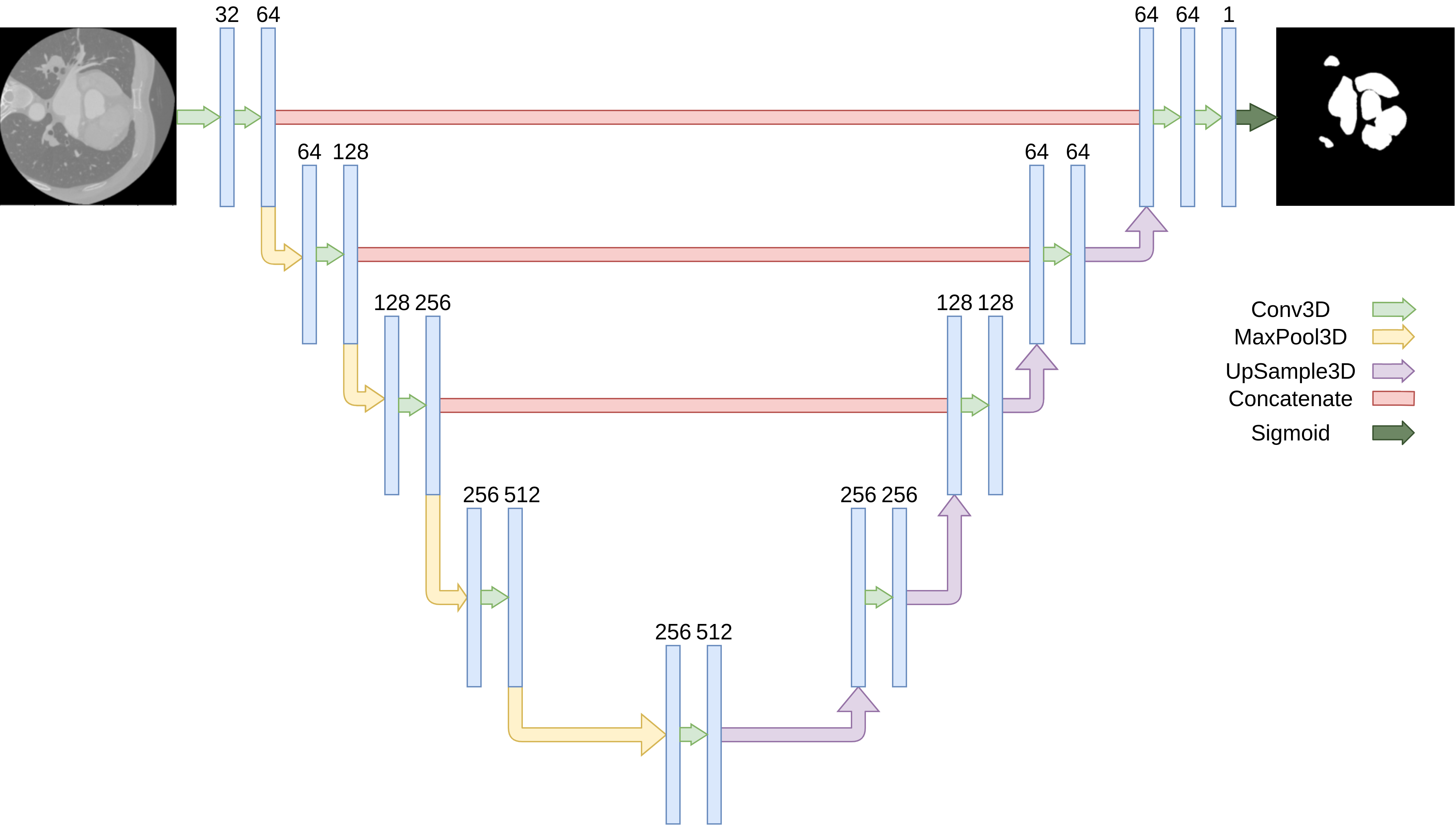
The Architecture contains 4 down
sampling steps with 2 consecutive Convolutions. All
convolutions used a RelU Activation function with filters that
are doubled at each convolution. The Training was done on a
Single GPU NVIDIA GTX 1080 TI with 11 GB Memory,
Python 3.6.7, CUDA 11.4 and libcudnn 8.2. Model Training
continued for about 100 epochs before reaching a plateau
where the Loss did not converge any more. The Data was
passed to the model after a series of Pre-Processing
Functions, Rescaling the data to the 0-1 Range, Applying
Resizing Kernels to fit the Input Scan Volumes into the
Model we resampled each volume to (112, 112, 112) and
Applied Different Augmentation Techniques, Random Axis
Flipping (First and Second Axes), Random Gamma
Corrections and Random Rotations in the Axial Direction of
±10 Degrees. The Loss Function used was Dice Loss and
Optimization done by Adam Optimizer with 0.0001 learning,
we applied a Decay callback on the Learning rate to decrease
the learning rate on plateau. Our U-Net implementation was
done in TensorFlow 2.5 using the Keras API.
A loadable module for 3D Slicer Platform written in python, with a flask server for easy deployment on cloud computing services.
Our main goal was to provide a tool that could automatically quantify calcium volume of the calcified plaques in the Coronary Artery.
This volume, also known as calcium score, is one of the risk factors used to predict the likelihood of Coronary Artery Disease Events occurring in the next few years, this process is currently being done manually by Radiologists, This takes a long time, and the results are subjective.
This is what motivated us to make this extension, as way to help make this process better and more streamlined as there is currently no widespread tool that helps on this front.
scikit-image tensorflow
Installed Automatically By The Extension In Slicer's Packaged Python
The Slicer Module SlicerProcesses is required and comes pre-packaged with the extension
flask scikit-image tensorflow pillow
A requirements file for easy setup is included and can be found here.
Our Main Module, The CaScoreModule has two main operation modes:
-
Local Mode: The data is processed locally on the user's machine, note that for optimal performance, a system capable of running TensorFlow Models is needed, A CUDA-Capable GPU will increase performance drastically, TensorFlow and other required packages needs to be installed inside Slicer's Packaged Python, A check will be done during first operation in this mode and all the required packages will be automatically installed.
-
Online Mode: The data is processed online, the volume is sent to a given URL which will process the data and send it back, an example server is included in this repo together with a requirements file.
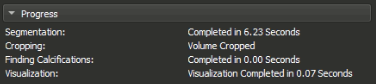
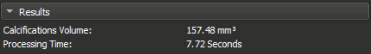
The Module's widget is divided into 6 main sections which helps the user in selecting the settings they need and also see the progress & the results. These sections are:

In this section, we simply select which one of the loaded volumes will we do the processing on

In this section, we select the main parameters for our processing:
We have two main modes in our module, a local mode and an online mode, This setting determines which one we will use, in summary the difference between them is:
-
Local Mode: The data is processed locally on the user's machine, note that for optimal performance, a system capable of running TensorFlow Models is needed, A CUDA-Capable GPU will increase performance drastically, TensorFlow and other required packages needs to be installed inside Slicer's Packaged Python, A check will be done during first operation in this mode and all the required packages will be automatically installed.
-
Online Mode: The data is processed online, the volume is sent to a given URL which will process the data and send it back, an example server is included in this repo together with a requirements file.
This setting enables cropping, it uses the results of a Deep Learning model built using TensorFlow to segment the heart, determining its location, then we create a bounding box around the ROI (The Heart) predicted by the model and crop out everything outside it.
This enables the Partial Segmentation mode, in this mode we select the three middle slices from the Axial, Sagittal & Coronal views and use them to get the bounding box's coordinates used in cropping, while it's not as accurate or useful as segmenting the whole volume, it's role shines as a pre-processing step, this rough cropping could be done before segmenting the whole volume to increase its quality, or decrease the volume's size which is useful in cases of using the online mode in a low or limited internet bandwidth setting.
Create a Segmentation node from the results of the full heart segmentation, enabling this disables the partial segmentation option, this segmentation is shown over the input heart CT image and shows the location of the heart, it could also be converted into a closed surface representation of the heart. This option is required to find calcifications.
Creates a closed surface representation of the heart's segmentation, this creates a 3D view of the heart which could have numerous benefits, this option requires the full volume segmentation and the creation of a segmentation node.
Uses one of two available methods to find calcifications in the heart and save it in a segmentation node, by default uses Image Processing techniques to determine the calcification the calculates their total volume. Requires full heart segmentation.
The image processing option uses image thresholding (>130-160) to find all locations containing calcium in the volume and saving it in a Calcifications volume, we then use it to get two different volume:
-
Calcifications in the heart's segmentation area, this is done by masking the calcifications using the heart segmentation prediction, so only calcification inside our predicted heart are detected.
-
Ignoring calcifications in the calcifications volume which are located near known bone masses.
By adding these two, we create a new volume which accurately locates all calcifications.
Creates a closed surface representation of the calcifications, similar to that of the heart.
Using this option utilizes a TensorFlow Deep Learning Model to predict the location of calcifications.
This option delegates long-running operation to a separate process so that it doesn't block 3D Slicer's UI.
This is a problem that plagues parts of 3D Slicer as it mainly works in a single-thread so any long-running or CPU intensive operations completely locks out the user from using the ui.
We choose to use a separate process instead of a second thread for a couple of reasons which are:
-
Using a different process for CPU-Intensive operations is generally preferred as it is faster than using threads, threads are better suited for I/O operations or any operation that simply involves waiting, since the main operation that we delegate is model prediction which could utilize the CPU heavily, running it in a separate process seemed more practical.
-
Due to the way 3D Slicer is built, and the fact that it runs on a pre-packaged python version and relies on it heavily to complete various operations (Even though 3D slicer is made using C++), threads are incompatible with it to a certain degree, to be able to use threads certain actions need to be made which could destabilize the application making it more error prone and prevent the usage of some functionalities temporarily, this was a point against using threads even though it's generally easier to use them inside Qt.

These settings are only available when using the local mode
Path to the folder containing the TensorFlow model used in the heart's segmentation, we have one already provided which can be downloaded, but any model would work.
Path to the folder containing the TensorFlow model used in detecting calcifications.
There is currently no available demo model with reasonable output for calcifications detection provided

These settings are only available when using the online mode.
URL of the server to send the data to for processing, a demo server is provided
Only available during the partial segmentation mode, returns the bounding box coordinates instead of returning the segmented slices and calculating the coordinates locally
Removes any possible patient personal information before sending the volume to the server, currently can't be disabled but this could change in the future when some information, such as age and sex, could be used as parameters to help more in diagnosis.
Shows the progress of the operations, what is currently in progress, what has been completed and some time statistics.
Shows the results of any calculation made, currently show the volume of the detected calcifications, also shows the total time taken to process the data.