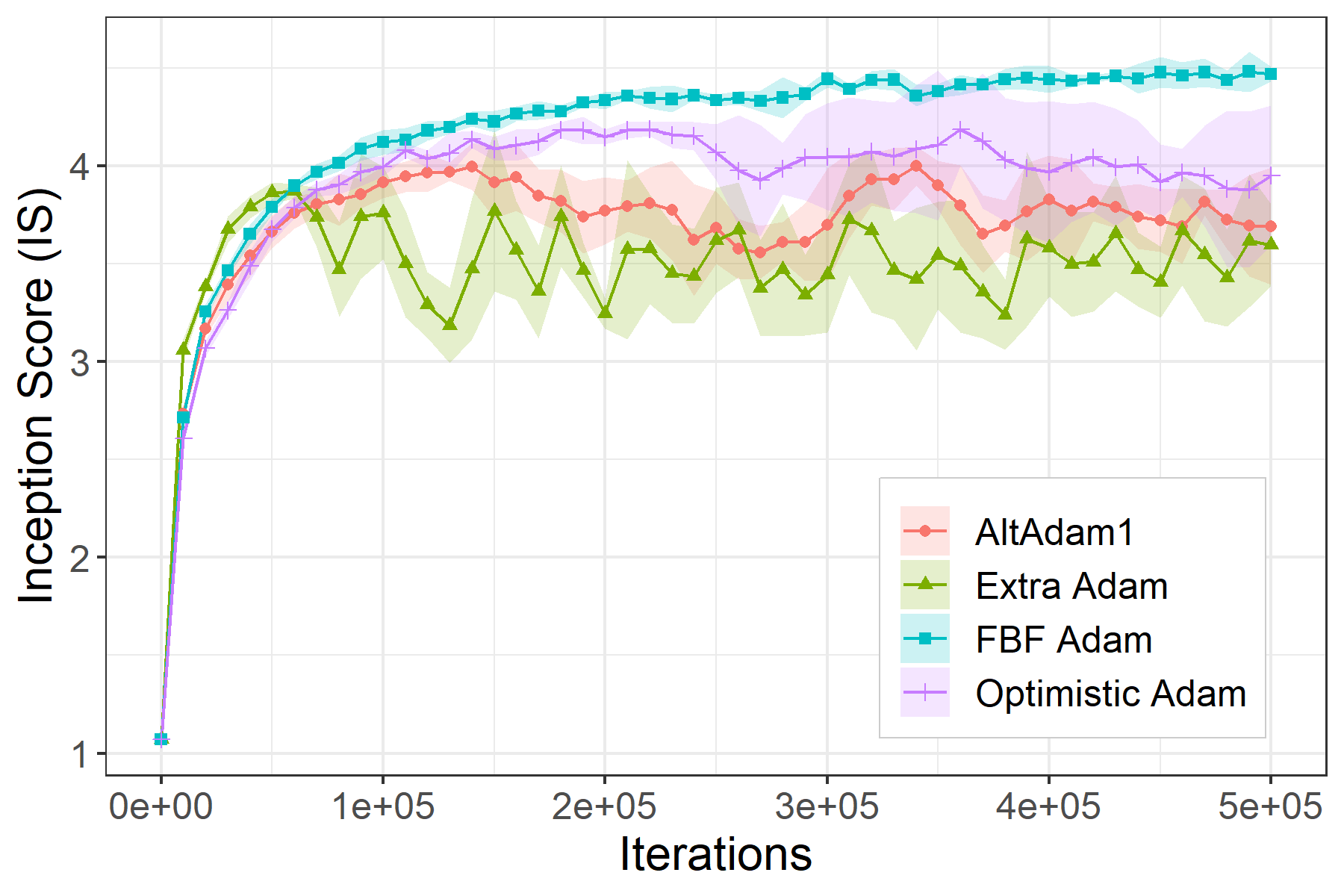
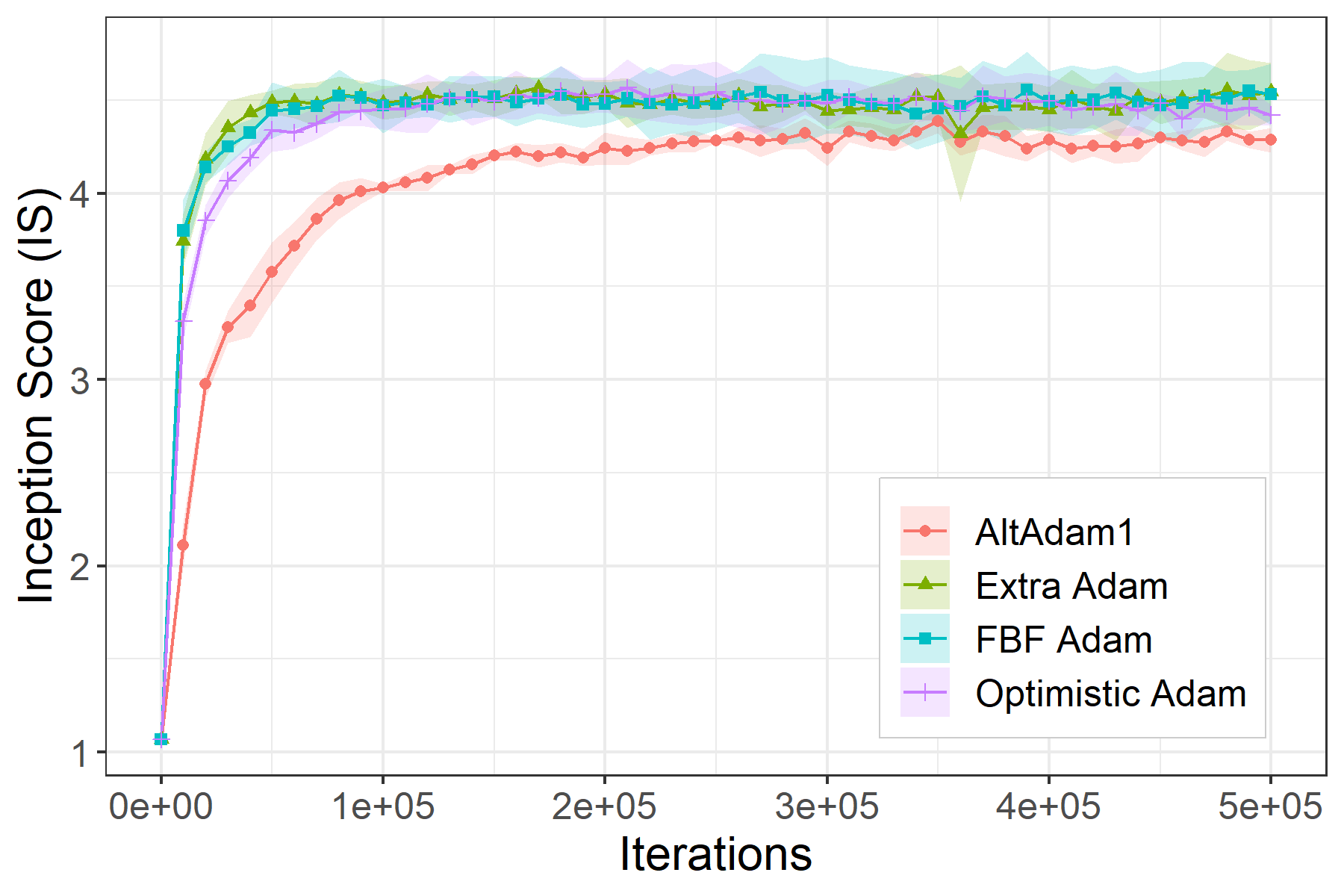
This is the code associated with the paper Two steps at a time --- taking GAN training in stride with Tseng's method. If you find this code useful please cite as:
Axel Böhm, Michael Sedlmayer, Robert Csetnek, Radu Bot Two steps at a time --- taking GAN training in stride with Tseng's method, 2020
The code base was mostly taken from here associated with the paper A Variational Inequality Perspective on Generative Adversarial Networks.
The code is in pytorch and was tested for:
- pytorch=1.3.1
(Optional) The inception score is computed using the updated implementation from A Note on the Inception Score which can be found here.
A conda environement is also provided (requires CUDA 10):
conda env create -f environment.yml
The Forward-Backward-Forward method is packaged as a torch.optim.Optimizer with an additional method extrapolation(). Two variants are available FBFSGD and FBFAdam.
Example of how to run FBF:
for i, input, target in enumerate(dataset):
FBF.zero_grad()
output = model(input)
loss = loss_fn(output, target)
loss.backward()
if i % 2:
FBF.extrapolation()
else:
FBF.step()To run the WGAN experiment with weight clipping using FBFAdam and the DCGAN architecture on CIFAR10 with the parameters from the paper:
python train_fbfadam.py output --default --cuda
To run the WGAN-L1 experiment with L1 regularization using FBFAdam and the DCGAN architecture on CIFAR10 with the parameters from the paper:
python train_fbfadam.py output --default --cuda -rp 0.0001
The --default option loads the hyperparameters used in the paper for each experiments, they are available as JSON files in the config folder.
The weights for our WGAN-L1 and DCGAN model trained with FBFAdam are available in the results folder.
For evaluation:
python eval_inception_score.py results/FBFAdam/best_DCGAN_WGAN-L1.state
and
python eval_fid.py results/FBFAdam/best_DCGAN_WGAN-L1.state
An ipython notebook is also available for the bilinear example here.