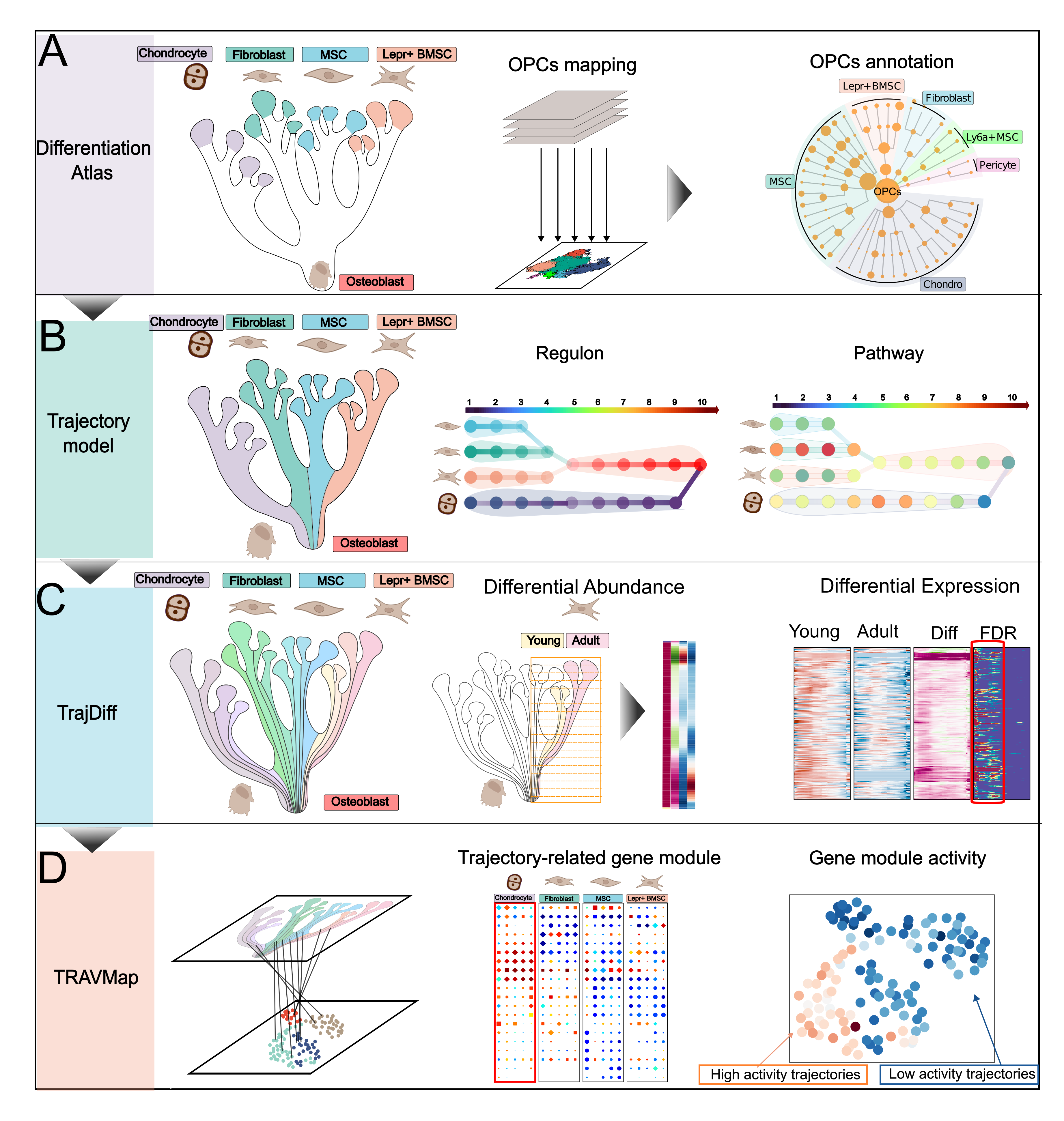
The central idea of TrajAtlas revolves around axis-based analysis. Although initially designed for osteogenesis datasets, it can be applied to almost any type of dataset as long as you have an "axis" (such as pseudotime or gene expression patterns).
Please refer to the documentation. In particular:
If you want to do pseudotime analysis, please check
Differential Abundance analysis
Differential Expression analysis
If you are interested in gene expression programs, we have developed a pipeline based on it. Please check
TrajAtlas meets gene expression program
If you are using gene set scoring for function inference, you can utilize TrajAtlas to identify which genes are more significant within the gene sets.
TrajAtlas meets gene set scoring
If you have osteogenesis datasets, you can project your datasets onto our model.
Projecting osteogenesis datasets
You need to have Python 3.8 or newer installed on your system. If you don't have Python installed, we recommend installing Mamba.
You can install TrajAtlas with following codes
pip install TrajAtlasTrajAtlas recently has posted on bioRxiv. If TrajAtlas helps you, please cite our work.