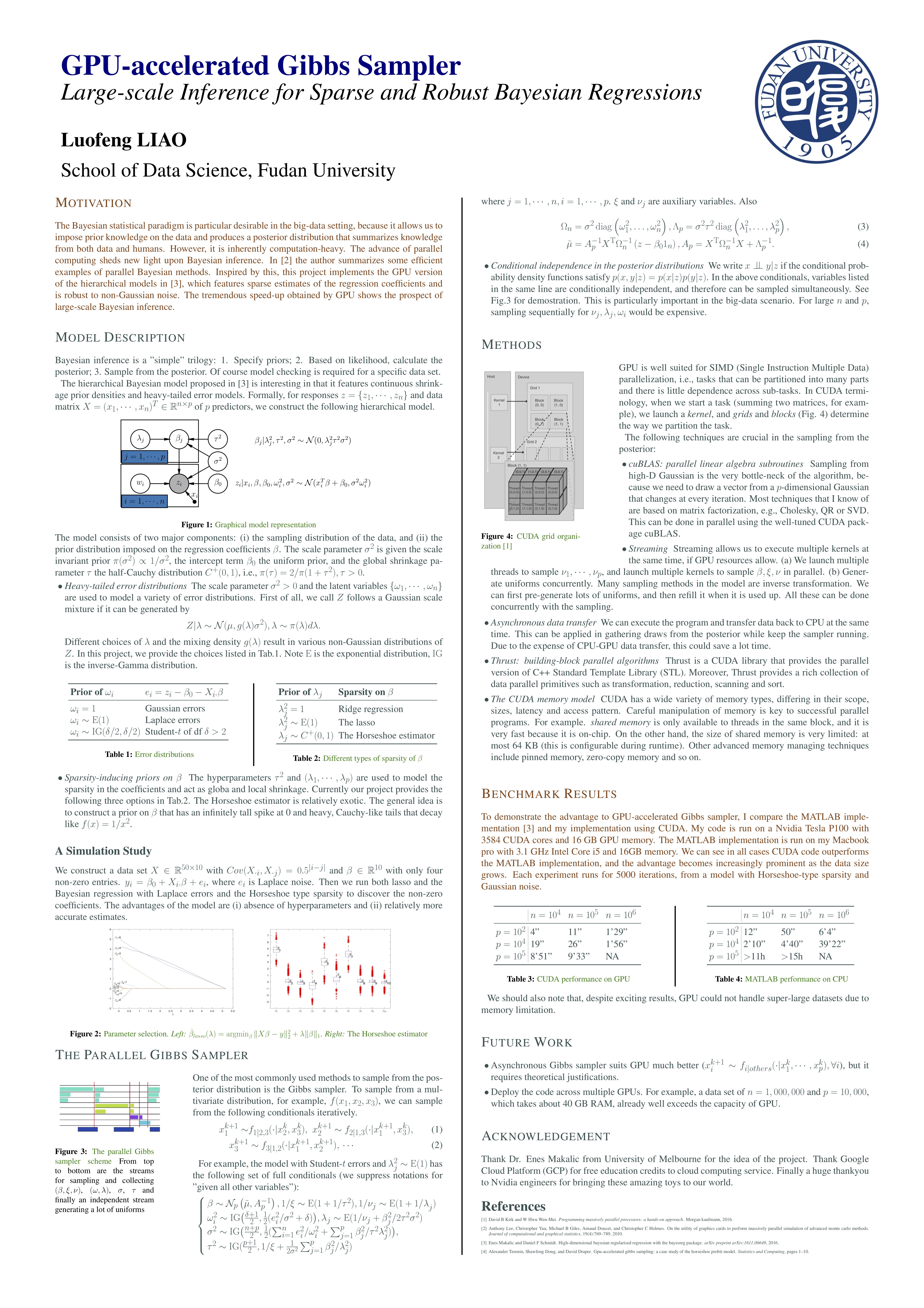
The poster here gives a thorough description of the entire project, including a description of the model, a simulation study comparing the model with lasso, and benchmark results in comparison with its MATLAB implementation. It gains at most 100x speed-up on super large dataset.
The poster is awarded the Runnerup Award out of around 30 equally competitive posters in the student poster section of 2018 Fudan Science and Innovation Forum.
This project implements the GPU version of the MATLAB Bayereg package [1], which features Bayesian linear regression with Gaussian or heavy-tailed error models with ridge, and lasso, horseshoe and horseshoe+ estimators
The package is developed under Ubuntu 16.0.
First, please make sure you have CUDA package installed. Check this by typing the following command.
nvcc --version
nvidia-smiIn my case, I have the following output.
$ nvcc --version
nvcc: NVIDIA (R) Cuda compiler driver
Copyright (c) 2005-2016 NVIDIA Corporation
Built on Tue_Jan_10_13:22:03_CST_2017
Cuda compilation tools, release 8.0, V8.0.61
$ nvidia-smi
Sat Dec 15 08:56:32 2018
+-----------------------------------------------------------------------------+
| NVIDIA-SMI 410.79 Driver Version: 410.79 CUDA Version: 10.0 |
|-------------------------------+----------------------+----------------------+
| GPU Name Persistence-M| Bus-Id Disp.A | Volatile Uncorr. ECC |
| Fan Temp Perf Pwr:Usage/Cap| Memory-Usage | GPU-Util Compute M. |
|===============================+======================+======================|
| 0 Tesla P100-PCIE... Off | 00000000:00:04.0 Off | 0 |
| N/A 40C P0 30W / 250W | 100MiB / 16280MiB | 0% Default |
+-------------------------------+----------------------+----------------------+
+-----------------------------------------------------------------------------+
| Processes: GPU Memory |
| GPU PID Type Process name Usage |
|=============================================================================|
| 0 1703 G /usr/lib/xorg/Xorg 100MiB |
+-----------------------------------------------------------------------------+Second, the tests in the program use R, so also make sure you install the following R pacakges. Make sure R can be called in any directory. (It's a little bit clumsy though.)
library("readr")
library("e1071")First, complie the package by typing make in the command line. To clean up the output files and the executable, type make clean.
By default, if you run ./a.out, the program will run a series of tests, using test files under the folder ./test_data. An example output should look like:
==== TEST 1: Initialization ====
[INFO]: Initialize a GpuGibbs object, data size: n = 4, p = 3.
[INFO]: Successful initialization
==== PASS TEST 1 ====
==== TEST 2: Residual update ====
[INFO]: Initialize a GpuGibbs object, data size: n = 4, p = 3.
[INFO]: Successful initialization
==== PASS TEST 2 ====
==== TEST 3: Gamma (single update, sigmaSqInv) dist ====
[INFO]: Initialize a GpuGibbs object, data size: n = 4, p = 3.
[INFO]: Successful initialization
Empirical estimate: alpha = 3.500000, beta = 2.000000
[1] "mean = 1.744453694125"
[1] "var = 0.874717715798537"
[1] "skew = 1.03556626743185"
Expected estimate:
[1] mean = 1.75
[1] var = 0.875
[1] skew = 1.069045
Empirical estimate: alpha = 3.500000, beta = 3.500000
[1] "mean = 1.00993304771613"
[1] "var = 0.289166113645945"
[1] "skew = 1.00366855371814"
...
./a.out 10000 1000 5000 0The above command will generate a dataset of 10000 x 1000, and run the Gibbs sampler for 5000 iterations. The 0 means we skip the testing. An example output should look like
$ ./a.out 10000 1000 5000 0
n=10000, p=1000, iter=5000
[INFO]: Initialize a GpuGibbs object, data size: n = 10000, p = 1000.
[INFO]: Start generating data
[INFO]: Finish generating data
[INFO]: Successful initialization
Iter = 0, time = 0.022564, expeted total = inf
Iter = 100, time = 0.426637, expeted total = 21.331847
Iter = 200, time = 0.810320, expeted total = 20.257998
Iter = 300, time = 1.190723, expeted total = 19.845382
Iter = 400, time = 1.572329, expeted total = 19.654110
Iter = 500, time = 1.950976, expeted total = 19.509759
Iter = 600, time = 2.325463, expeted total = 19.378857
...[1] Makalic, Enes, and Daniel F. Schmidt. "High-Dimensional Bayesian Regularised Regression with the BayesReg Package." arXiv preprint arXiv:1611.06649 (2016).
[2] Terenin, Alexander, Shawfeng Dong, and David Draper. "GPU-accelerated Gibbs sampling: a case study of the Horseshoe Probit model." Statistics and Computing 29.2 (2019): 301-310.